Lisanne van Dijk
DASS Good: Explainable Data Mining of Spatial Cohort Data
Apr 10, 2023Abstract:Developing applicable clinical machine learning models is a difficult task when the data includes spatial information, for example, radiation dose distributions across adjacent organs at risk. We describe the co-design of a modeling system, DASS, to support the hybrid human-machine development and validation of predictive models for estimating long-term toxicities related to radiotherapy doses in head and neck cancer patients. Developed in collaboration with domain experts in oncology and data mining, DASS incorporates human-in-the-loop visual steering, spatial data, and explainable AI to augment domain knowledge with automatic data mining. We demonstrate DASS with the development of two practical clinical stratification models and report feedback from domain experts. Finally, we describe the design lessons learned from this collaborative experience.
Slice-by-slice deep learning aided oropharyngeal cancer segmentation with adaptive thresholding for spatial uncertainty on FDG PET and CT images
Jul 04, 2022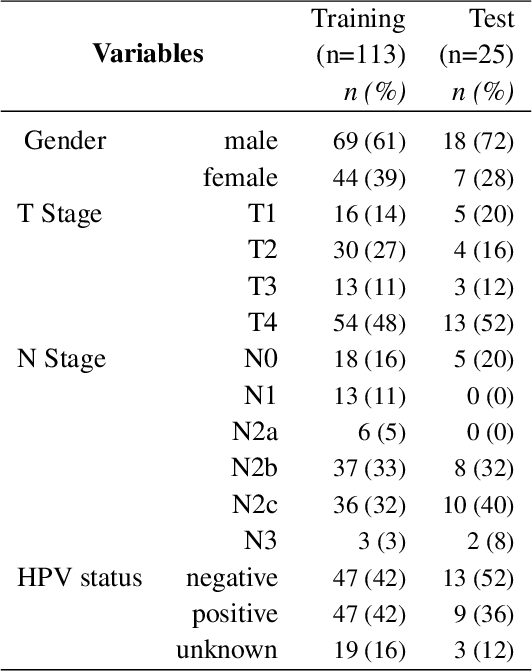
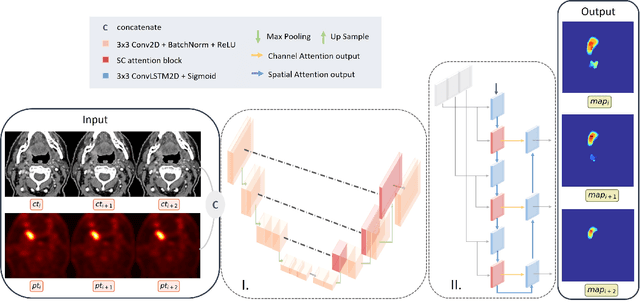
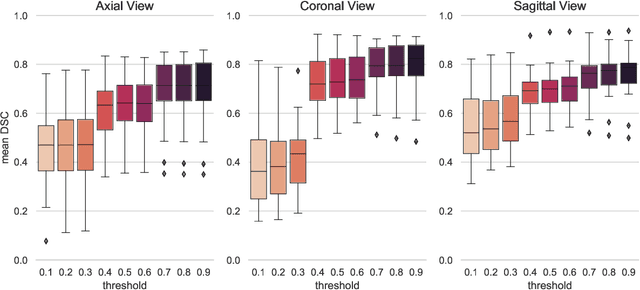
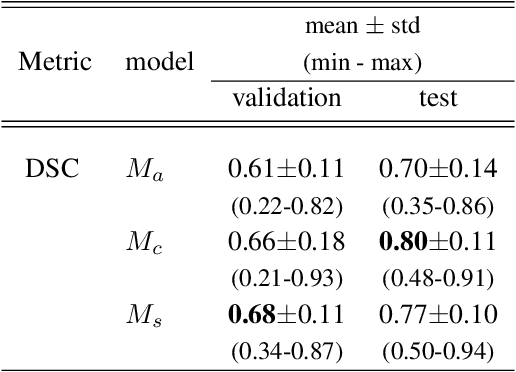
Abstract:Tumor segmentation is a fundamental step for radiotherapy treatment planning. To define an accurate segmentation of the primary tumor (GTVp) of oropharyngeal cancer patients (OPC), simultaneous assessment of different image modalities is needed, and each image volume is explored slice-by-slice from different orientations. Moreover, the manual fixed boundary of segmentation neglects the spatial uncertainty known to occur in tumor delineation. This study proposes a novel automatic deep learning (DL) model to assist radiation oncologists in a slice-by-slice adaptive GTVp segmentation on registered FDG PET/CT images. We included 138 OPC patients treated with (chemo)radiation in our institute. Our DL framework exploits both inter and intra-slice context. Sequences of 3 consecutive 2D slices of concatenated FDG PET/CT images and GTVp contours were used as input. A 3-fold cross validation was performed three times, training on sequences extracted from the Axial (A), Sagittal (S), and Coronal (C) plane of 113 patients. Since consecutive sequences in a volume contain overlapping slices, each slice resulted in three outcome predictions that were averaged. In the A, S, and C planes, the output shows areas with different probabilities of predicting the tumor. The performance of the models was assessed on 25 patients at different probability thresholds using the mean Dice Score Coefficient (DSC). Predictions were the closest to the ground truth at a probability threshold of 0.9 (DSC of 0.70 in the A, 0.77 in the S, and 0.80 in the C plane). The promising results of the proposed DL model show that the probability maps on registered FDG PET/CT images could guide radiation oncologists in a slice-by-slice adaptive GTVp segmentation.
Explainable Spatial Clustering: Leveraging Spatial Data in Radiation Oncology
Aug 25, 2020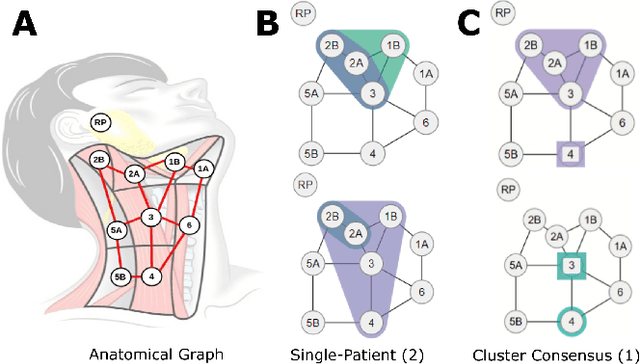
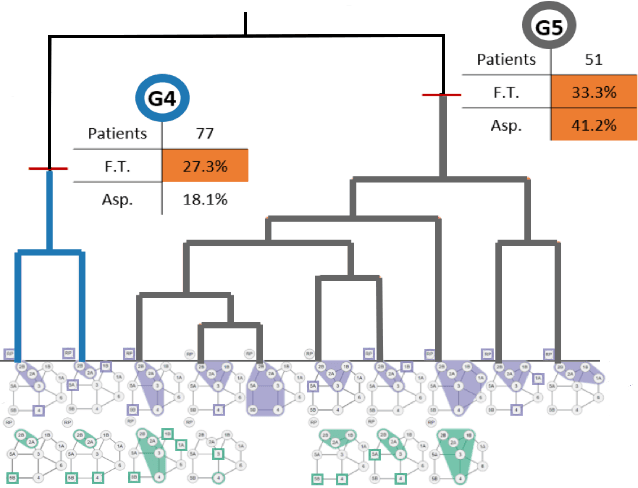
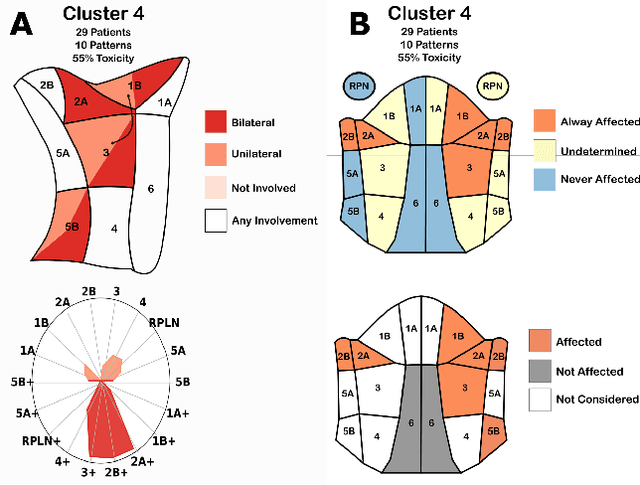
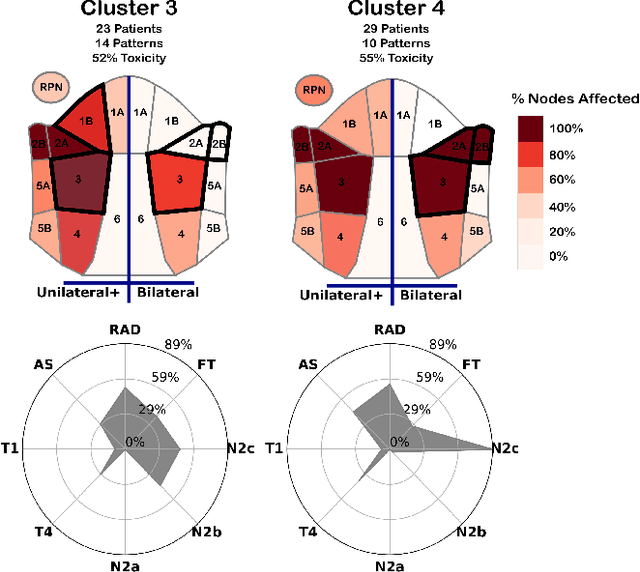
Abstract:Advances in data collection in radiation therapy have led to an abundance of opportunities for applying data mining and machine learning techniques to promote new data-driven insights. In light of these advances, supporting collaboration between machine learning experts and clinicians is important for facilitating better development and adoption of these models. Although many medical use-cases rely on spatial data, where understanding and visualizing the underlying structure of the data is important, little is known about the interpretability of spatial clustering results by clinical audiences. In this work, we reflect on the design of visualizations for explaining novel approaches to clustering complex anatomical data from head and neck cancer patients. These visualizations were developed, through participatory design, for clinical audiences during a multi-year collaboration with radiation oncologists and statisticians. We distill this collaboration into a set of lessons learned for creating visual and explainable spatial clustering for clinical users.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge