Lena Stempfle
Prediction Models That Learn to Avoid Missing Values
May 06, 2025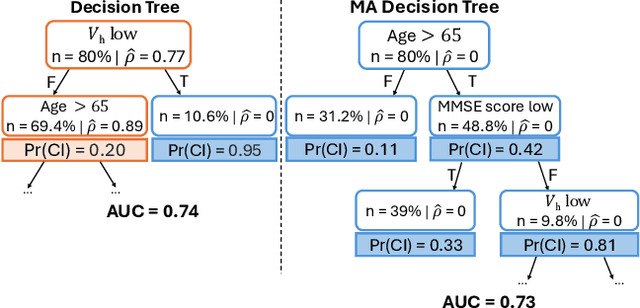
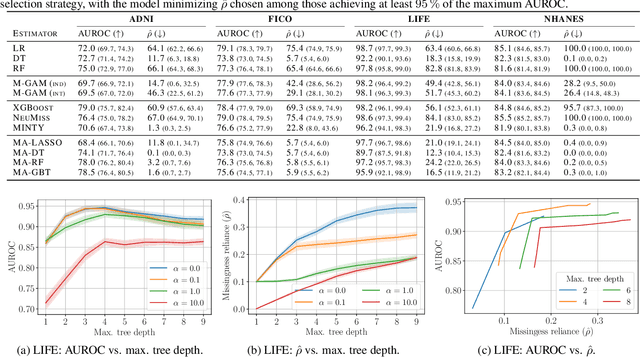
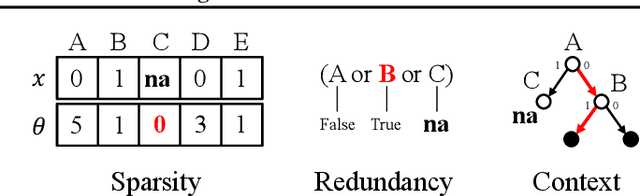

Abstract:Handling missing values at test time is challenging for machine learning models, especially when aiming for both high accuracy and interpretability. Established approaches often add bias through imputation or excessive model complexity via missingness indicators. Moreover, either method can obscure interpretability, making it harder to understand how the model utilizes the observed variables in predictions. We propose missingness-avoiding (MA) machine learning, a general framework for training models to rarely require the values of missing (or imputed) features at test time. We create tailored MA learning algorithms for decision trees, tree ensembles, and sparse linear models by incorporating classifier-specific regularization terms in their learning objectives. The tree-based models leverage contextual missingness by reducing reliance on missing values based on the observed context. Experiments on real-world datasets demonstrate that MA-DT, MA-LASSO, MA-RF, and MA-GBT effectively reduce the reliance on features with missing values while maintaining predictive performance competitive with their unregularized counterparts. This shows that our framework gives practitioners a powerful tool to maintain interpretability in predictions with test-time missing values.
How Should We Represent History in Interpretable Models of Clinical Policies?
Dec 10, 2024



Abstract:Modeling policies for sequential clinical decision-making based on observational data is useful for describing treatment practices, standardizing frequent patterns in treatment, and evaluating alternative policies. For each task, it is essential that the policy model is interpretable. Learning accurate models requires effectively capturing the state of a patient, either through sequence representation learning or carefully crafted summaries of their medical history. While recent work has favored the former, it remains a question as to how histories should best be represented for interpretable policy modeling. Focused on model fit, we systematically compare diverse approaches to summarizing patient history for interpretable modeling of clinical policies across four sequential decision-making tasks. We illustrate differences in the policies learned using various representations by breaking down evaluations by patient subgroups, critical states, and stages of treatment, highlighting challenges specific to common use cases. We find that interpretable sequence models using learned representations perform on par with black-box models across all tasks. Interpretable models using hand-crafted representations perform substantially worse when ignoring history entirely, but are made competitive by incorporating only a few aggregated and recent elements of patient history. The added benefits of using a richer representation are pronounced for subgroups and in specific use cases. This underscores the importance of evaluating policy models in the context of their intended use.
Expert Study on Interpretable Machine Learning Models with Missing Data
Nov 14, 2024



Abstract:Inherently interpretable machine learning (IML) models provide valuable insights for clinical decision-making but face challenges when features have missing values. Classical solutions like imputation or excluding incomplete records are often unsuitable in applications where values are missing at test time. In this work, we conducted a survey with 71 clinicians from 29 trauma centers across France, including 20 complete responses to study the interaction between medical professionals and IML applied to data with missing values. This provided valuable insights into how missing data is interpreted in clinical machine learning. We used the prediction of hemorrhagic shock as a concrete example to gauge the willingness and readiness of the participants to adopt IML models from three classes of methods. Our findings show that, while clinicians value interpretability and are familiar with common IML methods, classical imputation techniques often misalign with their intuition, and that models that natively handle missing values are preferred. These results emphasize the need to integrate clinical intuition into future IML models for better human-computer interaction.
MINTY: Rule-based Models that Minimize the Need for Imputing Features with Missing Values
Nov 23, 2023



Abstract:Rule models are often preferred in prediction tasks with tabular inputs as they can be easily interpreted using natural language and provide predictive performance on par with more complex models. However, most rule models' predictions are undefined or ambiguous when some inputs are missing, forcing users to rely on statistical imputation models or heuristics like zero imputation, undermining the interpretability of the models. In this work, we propose fitting concise yet precise rule models that learn to avoid relying on features with missing values and, therefore, limit their reliance on imputation at test time. We develop MINTY, a method that learns rules in the form of disjunctions between variables that act as replacements for each other when one or more is missing. This results in a sparse linear rule model, regularized to have small dependence on features with missing values, that allows a trade-off between goodness of fit, interpretability, and robustness to missing values at test time. We demonstrate the value of MINTY in experiments using synthetic and real-world data sets and find its predictive performance comparable or favorable to baselines, with smaller reliance on features with missing values.
Sharing pattern submodels for prediction with missing values
Jun 22, 2022



Abstract:Missing values are unavoidable in many applications of machine learning and present a challenge both during training and at test time. When variables are missing in recurring patterns, fitting separate pattern submodels have been proposed as a solution. However, independent models do not make efficient use of all available data. Conversely, fitting a shared model to the full data set typically relies on imputation which may be suboptimal when missingness depends on unobserved factors. We propose an alternative approach, called sharing pattern submodels, which make predictions that are a) robust to missing values at test time, b) maintains or improves the predictive power of pattern submodels, and c) has a short description enabling improved interpretability. We identify cases where sharing is provably optimal, even when missingness itself is predictive and when the prediction target depends on unobserved variables. Classification and regression experiments on synthetic data and two healthcare data sets demonstrate that our models achieve a favorable trade-off between pattern specialization and information sharing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge