Leif Hultin
MiceBoneChallenge: Micro-CT public dataset and six solutions for automatic growth plate detection in micro-CT mice bone scans
Nov 26, 2024



Abstract:Detecting and quantifying bone changes in micro-CT scans of rodents is a common task in preclinical drug development studies. However, this task is manual, time-consuming and subject to inter- and intra-observer variability. In 2024, Anonymous Company organized an internal challenge to develop models for automatic bone quantification. We prepared and annotated a high-quality dataset of 3D $\mu$CT bone scans from $83$ mice. The challenge attracted over $80$ AI scientists from around the globe who formed $23$ teams. The participants were tasked with developing a solution to identify the plane where the bone growth happens, which is essential for fully automatic segmentation of trabecular bone. As a result, six computer vision solutions were developed that can accurately identify the location of the growth plate plane. The solutions achieved the mean absolute error of $1.91\pm0.87$ planes from the ground truth on the test set, an accuracy level acceptable for practical use by a radiologist. The annotated 3D scans dataset along with the six solutions and source code, is being made public, providing researchers with opportunities to develop and benchmark their own approaches. The code, trained models, and the data will be shared.
Towards Fully Automated Segmentation of Rat Cardiac MRI by Leveraging Deep Learning Frameworks
Sep 09, 2021
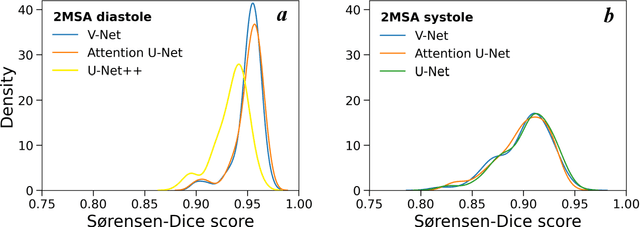


Abstract:Automated segmentation of human cardiac magnetic resonance datasets has been steadily improving during recent years. However, these methods are not directly applicable in preclinical context due to limited datasets and lower image resolution. Successful application of deep architectures for rat cardiac segmentation, although of critical importance for preclinical evaluation of cardiac function, has to our knowledge not yet been reported. We developed segmentation models that expand on the standard U-Net architecture and evaluated separate models for systole and diastole phases, 2MSA, and one model for all timepoints, 1MSA. Furthermore, we calibrated model outputs using a Gaussian Process (GP)-based prior to improve phase selection. Resulting models approach human performance in terms of left ventricular segmentation quality and ejection fraction (EF) estimation in both 1MSA and 2MSA settings (S{\o}rensen-Dice score 0.91 +/- 0.072 and 0.93 +/- 0.032, respectively). 2MSA achieved a mean absolute difference between estimated and reference EF of 3.5 +/- 2.5 %, while 1MSA resulted in 4.1 +/- 3.0 %. Applying Gaussian Processes to 1MSA allows to automate the selection of systole and diastole phases. Combined with a novel cardiac phase selection strategy, our work presents an important first step towards a fully automated segmentation pipeline in the context of rat cardiac analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge