Lang Zeng
Optimizing Cox Models with Stochastic Gradient Descent: Theoretical Foundations and Practical Guidances
Aug 05, 2024



Abstract:Optimizing Cox regression and its neural network variants poses substantial computational challenges in large-scale studies. Stochastic gradient descent (SGD), known for its scalability in model optimization, has recently been adapted to optimize Cox models. Unlike its conventional application, which typically targets a sum of independent individual loss, SGD for Cox models updates parameters based on the partial likelihood of a subset of data. Despite its empirical success, the theoretical foundation for optimizing Cox partial likelihood with SGD is largely underexplored. In this work, we demonstrate that the SGD estimator targets an objective function that is batch-size-dependent. We establish that the SGD estimator for the Cox neural network (Cox-NN) is consistent and achieves the optimal minimax convergence rate up to a polylogarithmic factor. For Cox regression, we further prove the $\sqrt{n}$-consistency and asymptotic normality of the SGD estimator, with variance depending on the batch size. Furthermore, we quantify the impact of batch size on Cox-NN training and its effect on the SGD estimator's asymptotic efficiency in Cox regression. These findings are validated by extensive numerical experiments and provide guidance for selecting batch sizes in SGD applications. Finally, we demonstrate the effectiveness of SGD in a real-world application where GD is unfeasible due to the large scale of data.
Dynamic Prediction using Time-Dependent Cox Survival Neural Network
Jul 12, 2023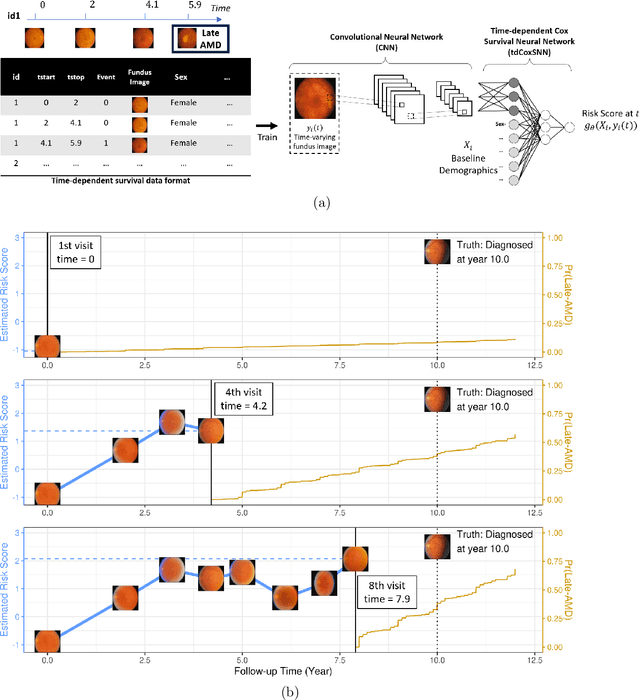
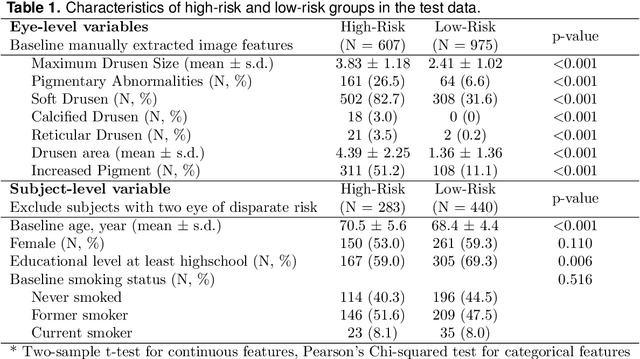
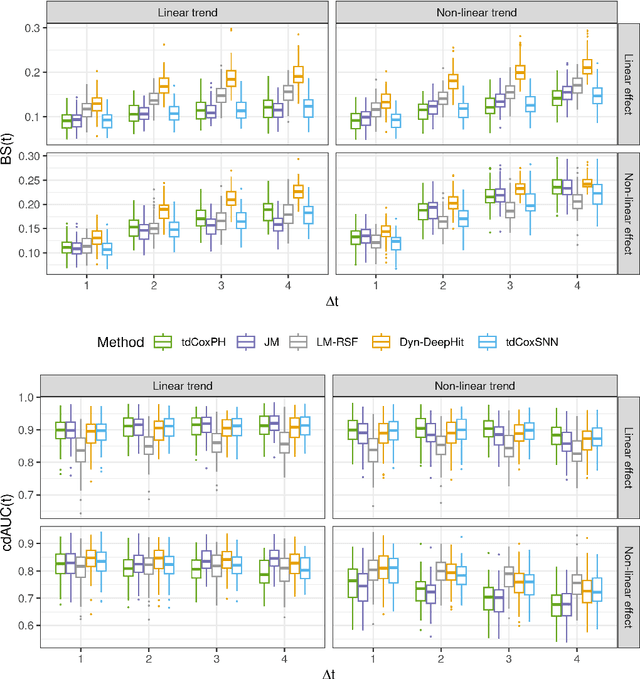
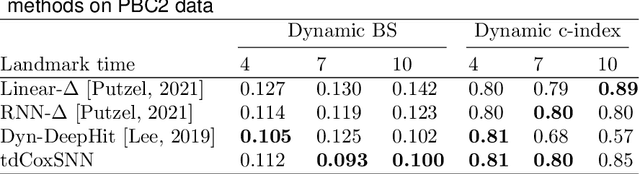
Abstract:The target of dynamic prediction is to provide individualized risk predictions over time which can be updated as new data become available. Motivated by establishing a dynamic prediction model for the progressive eye disease, age-related macular degeneration (AMD), we proposed a time-dependent Cox model-based survival neural network (tdCoxSNN) to predict its progression on a continuous time scale using longitudinal fundus images. tdCoxSNN extends the time-dependent Cox model by utilizing a neural network to model the non-linear effect of the time-dependent covariates on the survival outcome. Additionally, by incorporating the convolutional neural network (CNN), tdCoxSNN can take the longitudinal raw images as input. We evaluate and compare our proposed method with joint modeling and landmarking approaches through comprehensive simulations using two time-dependent accuracy metrics, the Brier Score and dynamic AUC. We applied the proposed approach to two real datasets. One is a large AMD study, the Age-Related Eye Disease Study (AREDS), in which more than 50,000 fundus images were captured over a period of 12 years for more than 4,000 participants. Another is a public dataset of the primary biliary cirrhosis (PBC) disease, in which multiple lab tests were longitudinally collected to predict the time-to-liver transplant. Our approach achieves satisfactory prediction performance in both simulation studies and the two real data analyses. tdCoxSNN was implemented in PyTorch, Tensorflow, and R-Tensorflow.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge