Kup-sze Choi
Generative Fuzzy System for Sequence Generation
Nov 21, 2024



Abstract:Generative Models (GMs), particularly Large Language Models (LLMs), have garnered significant attention in machine learning and artificial intelligence for their ability to generate new data by learning the statistical properties of training data and creating data that resemble the original. This capability offers a wide range of applications across various domains. However, the complex structures and numerous model parameters of GMs make the input-output processes opaque, complicating the understanding and control of outputs. Moreover, the purely data-driven learning mechanism limits GM's ability to acquire broader knowledge. There remains substantial potential for enhancing the robustness and generalization capabilities of GMs. In this work, we introduce the fuzzy system, a classical modeling method that combines data and knowledge-driven mechanisms, to generative tasks. We propose a novel Generative Fuzzy System framework, named GenFS, which integrates the deep learning capabilities of GM with the interpretability and dual-driven mechanisms of fuzzy systems. Specifically, we propose an end-to-end GenFS-based model for sequence generation, called FuzzyS2S. A series of experimental studies were conducted on 12 datasets, covering three distinct categories of generative tasks: machine translation, code generation, and summary generation. The results demonstrate that FuzzyS2S outperforms the Transformer in terms of accuracy and fluency. Furthermore, it exhibits better performance on some datasets compared to state-of-the-art models T5 and CodeT5.
Cooperation Learning Enhanced Colonic Polyp Segmentation Based on Transformer-CNN Fusion
Jan 17, 2023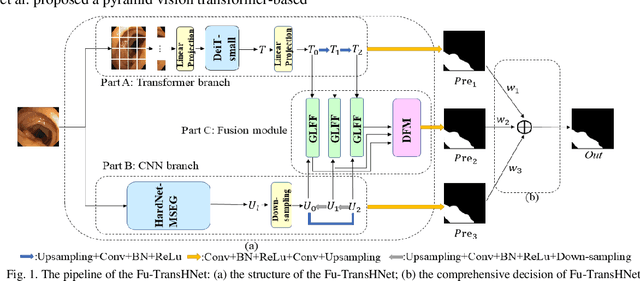
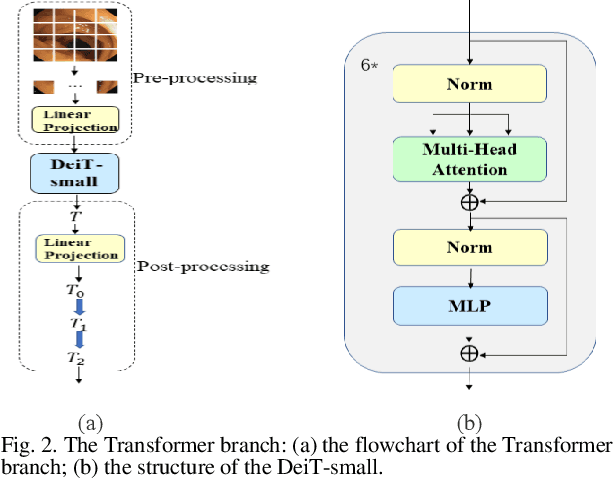
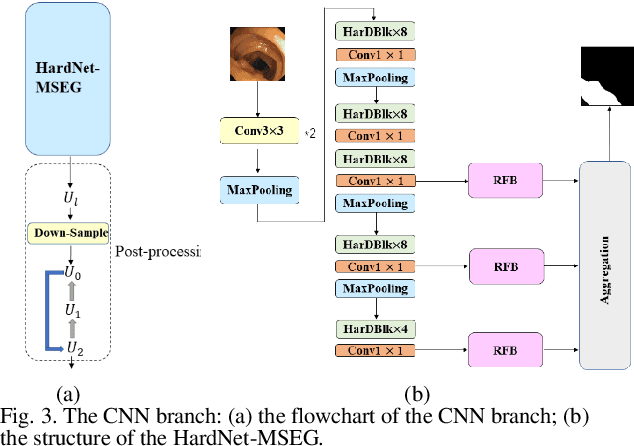
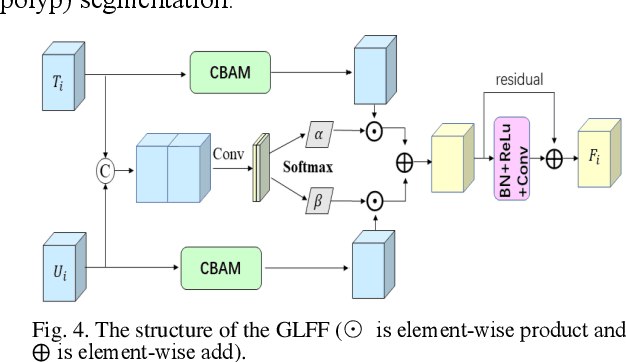
Abstract:Traditional segmentation methods for colonic polyps are mainly designed based on low-level features. They could not accurately extract the location of small colonic polyps. Although the existing deep learning methods can improve the segmentation accuracy, their effects are still unsatisfied. To meet the above challenges, we propose a hybrid network called Fusion-Transformer-HardNetMSEG (i.e., Fu-TransHNet) in this study. Fu-TransHNet uses deep learning of different mechanisms to fuse each other and is enhanced with multi-view collaborative learning techniques. Firstly, the Fu-TransHNet utilizes the Transformer branch and the CNN branch to realize the global feature learning and local feature learning, respectively. Secondly, a fusion module is designed to integrate the features from two branches. The fusion module consists of two parts: 1) the Global-Local Feature Fusion (GLFF) part and 2) the Dense Fusion of Multi-scale features (DFM) part. The former is built to compensate the feature information mission from two branches at the same scale; the latter is constructed to enhance the feature representation. Thirdly, the above two branches and fusion modules utilize multi-view cooperative learning techniques to obtain their respective weights that denote their importance and then make a final decision comprehensively. Experimental results showed that the Fu-TransHNet network was superior to the existing methods on five widely used benchmark datasets. In particular, on the ETIS-LaribPolypDB dataset containing many small-target colonic polyps, the mDice obtained by Fu-TransHNet were 12.4% and 6.2% higher than the state-of-the-art methods HardNet-MSEG and TransFuse-s, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge