Konstantinos Pliakos
HypeRS: Building a Hypergraph-driven ensemble Recommender System
Jun 22, 2023Abstract:Recommender systems are designed to predict user preferences over collections of items. These systems process users' previous interactions to decide which items should be ranked higher to satisfy their desires. An ensemble recommender system can achieve great recommendation performance by effectively combining the decisions generated by individual models. In this paper, we propose a novel ensemble recommender system that combines predictions made by different models into a unified hypergraph ranking framework. This is the first time that hypergraph ranking has been employed to model an ensemble of recommender systems. Hypergraphs are generalizations of graphs where multiple vertices can be connected via hyperedges, efficiently modeling high-order relations. We differentiate real and predicted connections between users and items by assigning different hyperedge weights to individual recommender systems. We perform experiments using four datasets from the fields of movie, music and news media recommendation. The obtained results show that the ensemble hypergraph ranking method generates more accurate recommendations compared to the individual models and a weighted hybrid approach. The assignment of different hyperedge weights to the ensemble hypergraph further improves the performance compared to a setting with identical hyperedge weights.
Explaining random forest prediction through diverse rulesets
Mar 29, 2022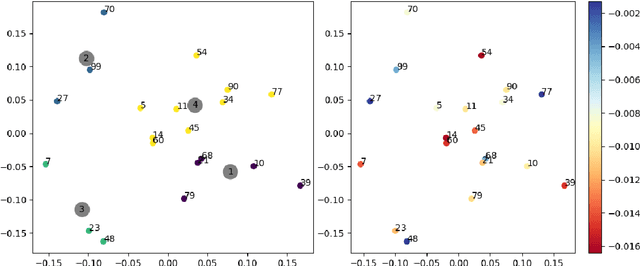

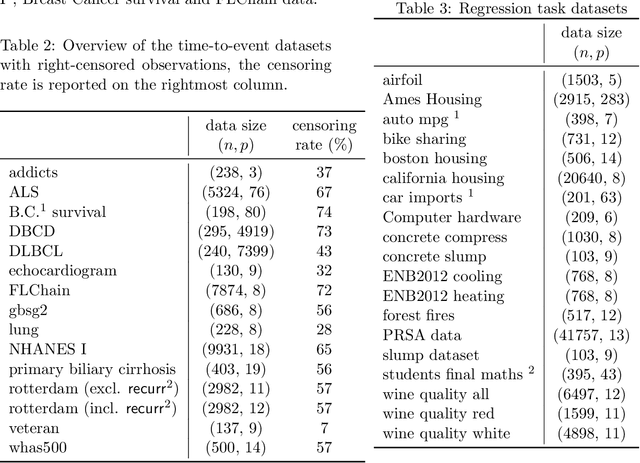
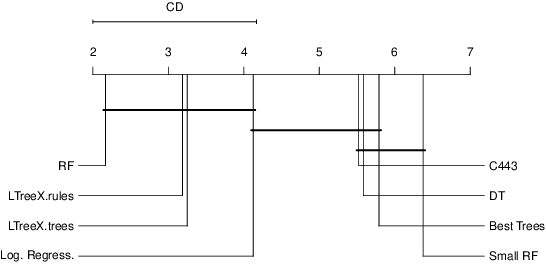
Abstract:Tree-ensemble algorithms, such as random forest, are effective machine learning methods popular for their flexibility, high performance, and robustness to overfitting. However, since multiple learners are combined,they are not as interpretable as a single decision tree. In this work we propose a methodology, called Local Tree eXtractor (LTreeX) which is able to explain the forest prediction for a given test instance with a few diverse rules. Starting from the decision trees generated by a random forest, our method 1) pre-selects a subset of them, 2) creates a vector representation, and 3) eventually clusters such a representation. Each cluster prototype results in a rule that explains the test instance prediction. We test the effectiveness of LTreeX on 71 real-world datasets and we demonstrate the validity of our approach for binary classification, regression, multi-label classification and time-to-event tasks. In all set-ups, we show that our extracted surrogate model manages to approximate the performance of the corresponding ensemble model, while selecting only few trees from the whole forest.We also show that our proposed approach substantially outperforms other explainable methods in terms of predictive performance.
Drug-Target Interaction Prediction via an Ensemble of Weighted Nearest Neighbors with Interaction Recovery
Dec 22, 2020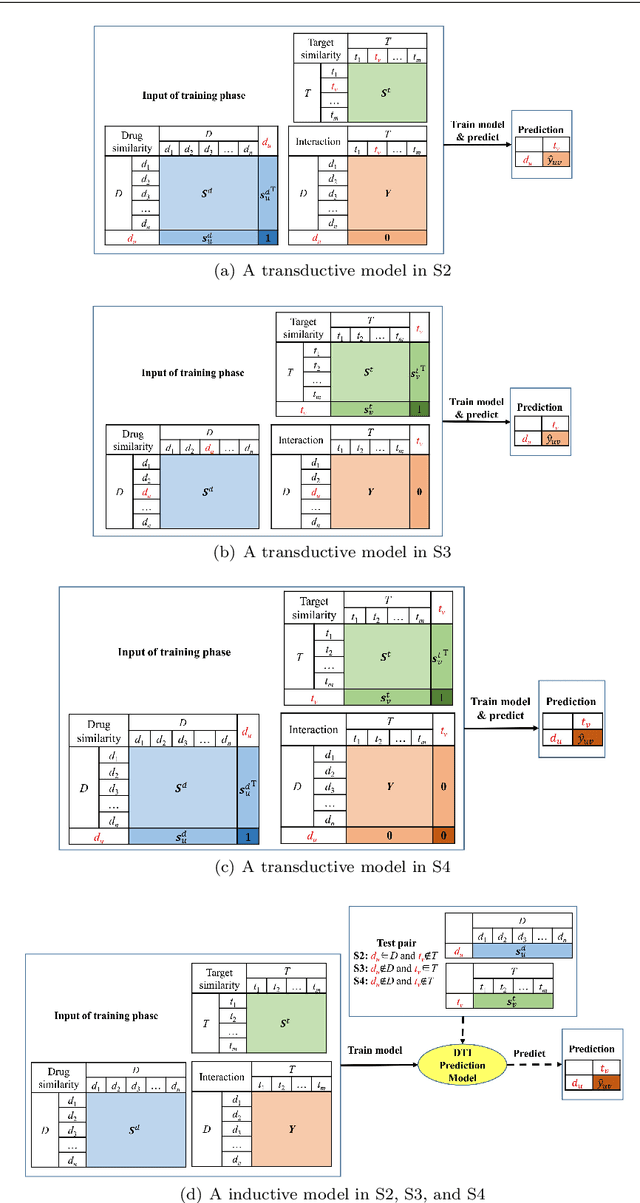
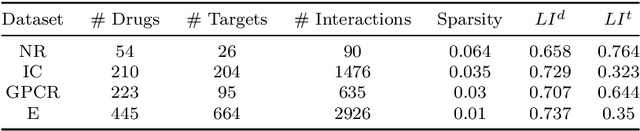
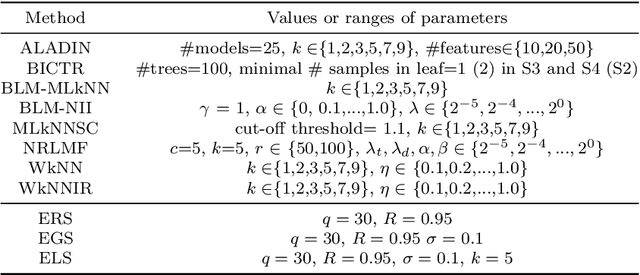

Abstract:Predicting drug-target interactions (DTI) via reliable computational methods is an effective and efficient way to mitigate the enormous costs and time of the drug discovery process. Structure-based drug similarities and sequence-based target protein similarities are the commonly used information for DTI prediction. Among numerous computational methods, neighborhood-based chemogenomic approaches that leverage drug and target similarities to perform predictions directly are simple but promising ones. However, most existing similarity-based methods follow the transductive setting. These methods cannot directly generalize to unseen data because they should be re-built to predict the interactions for new arriving drugs, targets, or drug-target pairs. Besides, many similarity-based methods, especially neighborhood-based ones, cannot handle directly all three types of interaction prediction. Furthermore, a large amount of missing interactions in current DTI datasets hinders most DTI prediction methods. To address these issues, we propose a new method denoted as Weighted k Nearest Neighbor with Interaction Recovery (WkNNIR). Not only can WkNNIR estimate interactions of any new drugs and/or new targets without any need of re-training, but it can also recover missing interactions. In addition, WkNNIR exploits local imbalance to promote the influence of more reliable similarities on the DTI prediction process. We also propose a series of ensemble methods that employ diverse sampling strategies and could be coupled with WkNNIR as well as any other DTI prediction method to improve performance. Experimental results over four benchmark datasets demonstrate the effectiveness of our approaches in predicting drug-target interactions. Lastly, we confirm the practical prediction ability of proposed methods to discover reliable interactions that not reported in the original benchmark datasets.
Fair Multi-Stakeholder News Recommender System with Hypergraph ranking
Dec 01, 2020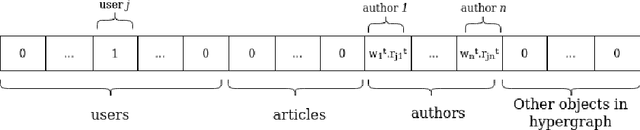

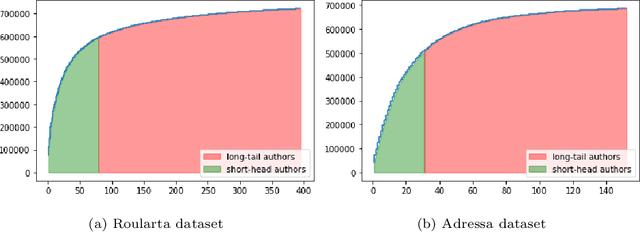

Abstract:Recommender systems are typically designed to fulfill end user needs. However, in some domains the users are not the only stakeholders in the system. For instance, in a news aggregator website users, authors, magazines as well as the platform itself are potential stakeholders. Most of the collaborative filtering recommender systems suffer from popularity bias. Therefore, if the recommender system only considers users' preferences, presumably it over-represents popular providers and under-represents less popular providers. To address this issue one should consider other stakeholders in the generated ranked lists. In this paper we demonstrate that hypergraph learning has the natural capability of handling a multi-stakeholder recommendation task. A hypergraph can model high order relations between different types of objects and therefore is naturally inclined to generate recommendation lists considering multiple stakeholders. We form the recommendations in time-wise rounds and learn to adapt the weights of stakeholders to increase the coverage of low-covered stakeholders over time. The results show that the proposed approach counters popularity bias and produces fairer recommendations with respect to authors in two news datasets, at a low cost in precision.
Deep tree-ensembles for multi-output prediction
Nov 03, 2020
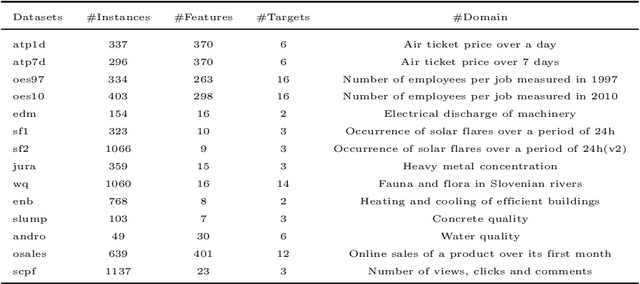

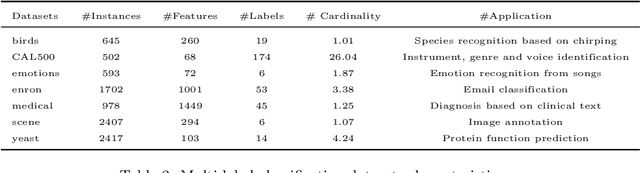
Abstract:Recently, deep neural networks have expanded the state-of-art in various scientific fields and provided solutions to long standing problems across multiple application domains. Nevertheless, they also suffer from weaknesses since their optimal performance depends on massive amounts of training data and the tuning of an extended number of parameters. As a countermeasure, some deep-forest methods have been recently proposed, as efficient and low-scale solutions. Despite that, these approaches simply employ label classification probabilities as induced features and primarily focus on traditional classification and regression tasks, leaving multi-output prediction under-explored. Moreover, recent work has demonstrated that tree-embeddings are highly representative, especially in structured output prediction. In this direction, we propose a novel deep tree-ensemble (DTE) model, where every layer enriches the original feature set with a representation learning component based on tree-embeddings. In this paper, we specifically focus on two structured output prediction tasks, namely multi-label classification and multi-target regression. We conducted experiments using multiple benchmark datasets and the obtained results confirm that our method provides superior results to state-of-the-art methods in both tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge