Drug-Target Interaction Prediction via an Ensemble of Weighted Nearest Neighbors with Interaction Recovery
Paper and Code
Dec 22, 2020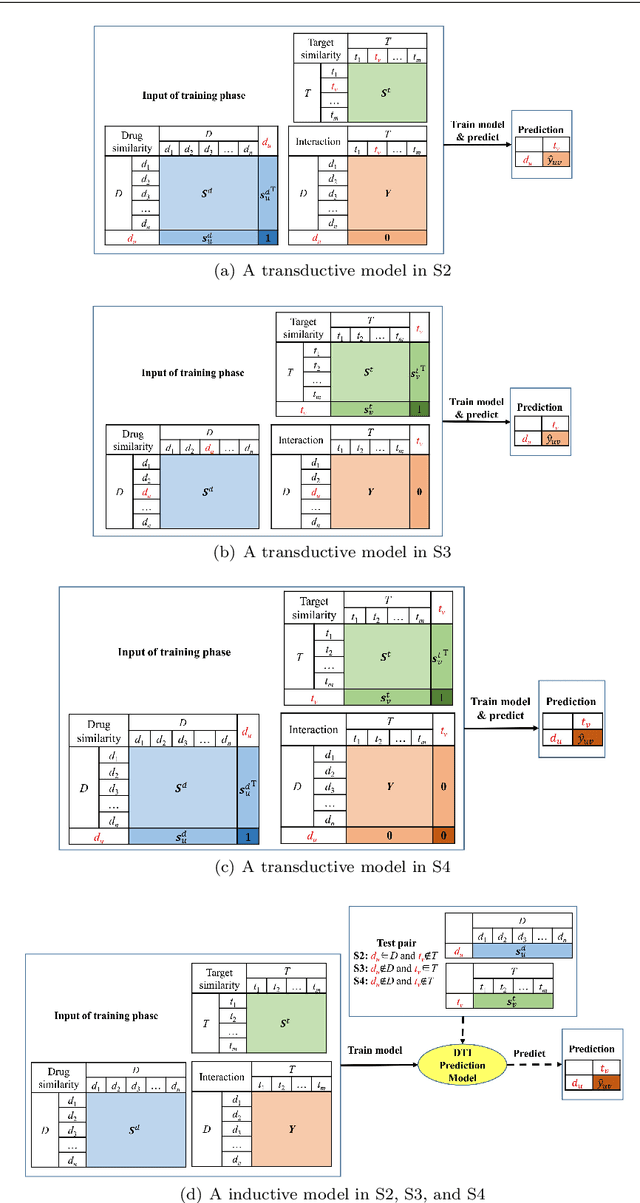
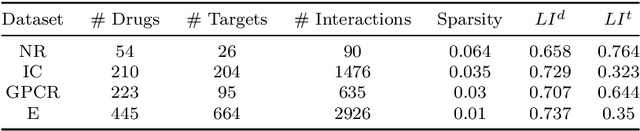
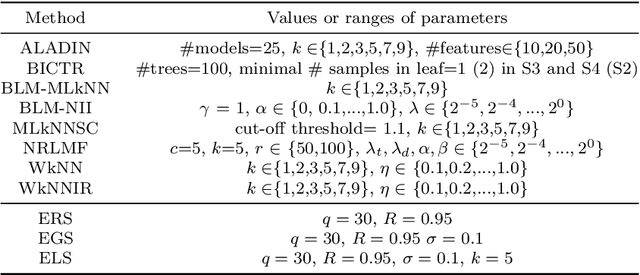

Predicting drug-target interactions (DTI) via reliable computational methods is an effective and efficient way to mitigate the enormous costs and time of the drug discovery process. Structure-based drug similarities and sequence-based target protein similarities are the commonly used information for DTI prediction. Among numerous computational methods, neighborhood-based chemogenomic approaches that leverage drug and target similarities to perform predictions directly are simple but promising ones. However, most existing similarity-based methods follow the transductive setting. These methods cannot directly generalize to unseen data because they should be re-built to predict the interactions for new arriving drugs, targets, or drug-target pairs. Besides, many similarity-based methods, especially neighborhood-based ones, cannot handle directly all three types of interaction prediction. Furthermore, a large amount of missing interactions in current DTI datasets hinders most DTI prediction methods. To address these issues, we propose a new method denoted as Weighted k Nearest Neighbor with Interaction Recovery (WkNNIR). Not only can WkNNIR estimate interactions of any new drugs and/or new targets without any need of re-training, but it can also recover missing interactions. In addition, WkNNIR exploits local imbalance to promote the influence of more reliable similarities on the DTI prediction process. We also propose a series of ensemble methods that employ diverse sampling strategies and could be coupled with WkNNIR as well as any other DTI prediction method to improve performance. Experimental results over four benchmark datasets demonstrate the effectiveness of our approaches in predicting drug-target interactions. Lastly, we confirm the practical prediction ability of proposed methods to discover reliable interactions that not reported in the original benchmark datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge