Kianoush Falahkheirkhah
Finer Disentanglement of Aleatoric Uncertainty Can Accelerate Chemical Histopathology Imaging
Feb 27, 2025Abstract:Label-free chemical imaging holds significant promise for improving digital pathology workflows. However, data acquisition speed remains a limiting factor for smooth clinical transition. To address this gap, we propose an adaptive strategy: initially scan the low information (LI) content of the entire tissue quickly, identify regions with high aleatoric uncertainty (AU), and selectively re-image them at better quality to capture higher information (HI) details. The primary challenge lies in distinguishing between high-AU regions that can be mitigated through HI imaging and those that cannot. However, since existing uncertainty frameworks cannot separate such AU subcategories, we propose a fine-grained disentanglement method based on post-hoc latent space analysis to unmix resolvable from irresolvable high-AU regions. We apply our approach to efficiently image infrared spectroscopic data of breast tissues, achieving superior segmentation performance using the acquired HI data compared to a random baseline. This represents the first algorithmic study focused on fine-grained AU disentanglement within dynamic image spaces (LI-to-HI), with novel application to streamline histopathology.
Detecting Hallucinations in Virtual Histology with Neural Precursors
Nov 22, 2024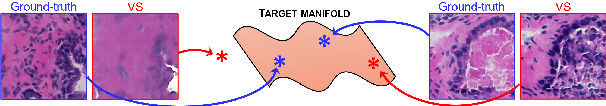
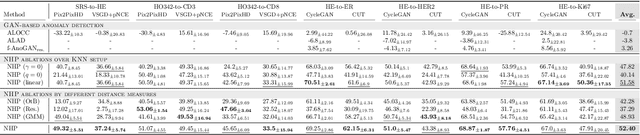
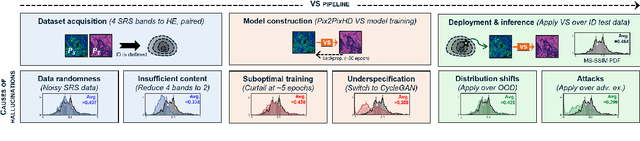
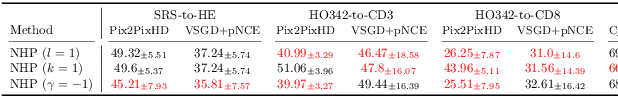
Abstract:Significant biomedical research and clinical care rely on the histopathologic examination of tissue structure using microscopy of stained tissue. Virtual staining (VS) offers a promising alternative with the potential to reduce cost and eliminate the use of toxic reagents. However, the critical challenge of hallucinations limits confidence in its use, necessitating a VS co-pilot to detect these hallucinations. Here, we first formally establish the problem of hallucination detection in VS. Next, we introduce a scalable, post-hoc hallucination detection method that identifies a Neural Hallucination Precursor (NHP) from VS model embeddings for test-time detection. We report extensive validation across diverse and challenging VS settings to demonstrate NHP's effectiveness and robustness. Furthermore, we show that VS models with fewer hallucinations do not necessarily disclose them better, risking a false sense of security when reporting just the former metric. This highlights the need for a reassessment of current VS evaluation practices.
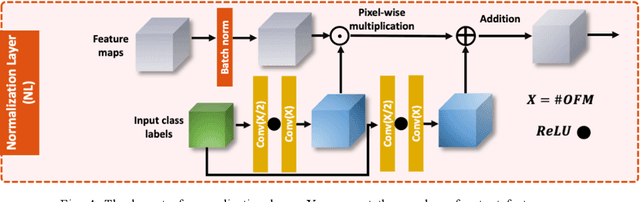
Are We Ready for Out-of-Distribution Detection in Digital Pathology?
Jul 18, 2024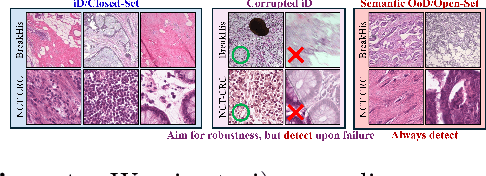

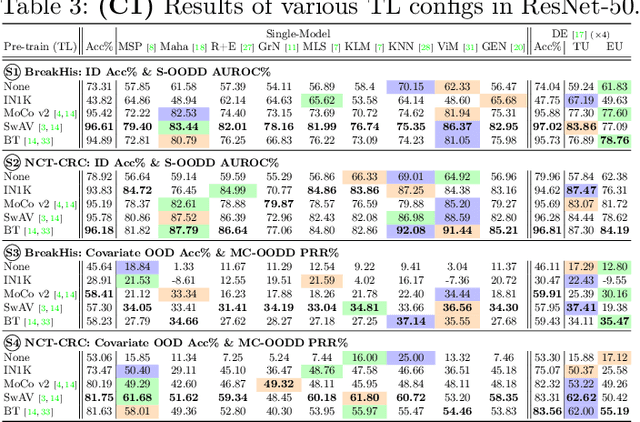
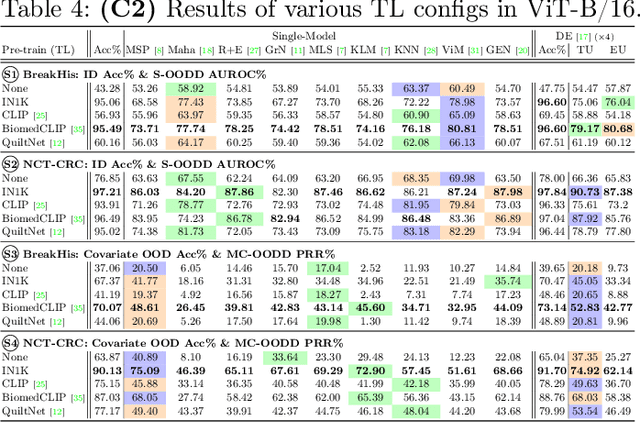
Abstract:The detection of semantic and covariate out-of-distribution (OOD) examples is a critical yet overlooked challenge in digital pathology (DP). Recently, substantial insight and methods on OOD detection were presented by the ML community, but how do they fare in DP applications? To this end, we establish a benchmark study, our highlights being: 1) the adoption of proper evaluation protocols, 2) the comparison of diverse detectors in both a single and multi-model setting, and 3) the exploration into advanced ML settings like transfer learning (ImageNet vs. DP pre-training) and choice of architecture (CNNs vs. transformers). Through our comprehensive experiments, we contribute new insights and guidelines, paving the way for future research and discussion.
Domain adaptation using optimal transport for invariant learning using histopathology datasets
Mar 03, 2023Abstract:Histopathology is critical for the diagnosis of many diseases, including cancer. These protocols typically require pathologists to manually evaluate slides under a microscope, which is time-consuming and subjective, leading to interest in machine learning to automate analysis. However, computational techniques are limited by batch effects, where technical factors like differences in preparation protocol or scanners can alter the appearance of slides, causing models trained on one institution to fail when generalizing to others. Here, we propose a domain adaptation method that improves the generalization of histopathological models to data from unseen institutions, without the need for labels or retraining in these new settings. Our approach introduces an optimal transport (OT) loss, that extends adversarial methods that penalize models if images from different institutions can be distinguished in their representation space. Unlike previous methods, which operate on single samples, our loss accounts for distributional differences between batches of images. We show that on the Camelyon17 dataset, while both methods can adapt to global differences in color distribution, only our OT loss can reliably classify a cancer phenotype unseen during training. Together, our results suggest that OT improves generalization on rare but critical phenotypes that may only make up a small fraction of the total tiles and variation in a slide.
Deepfake histological images for enhancing digital pathology
Jun 16, 2022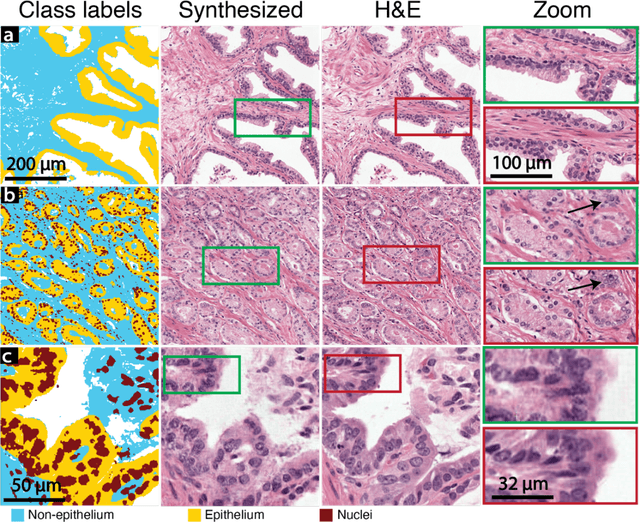
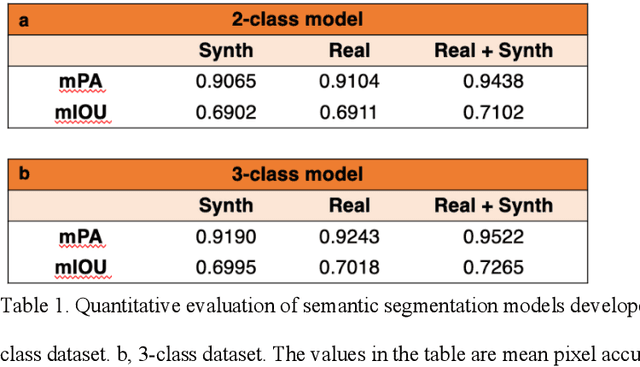
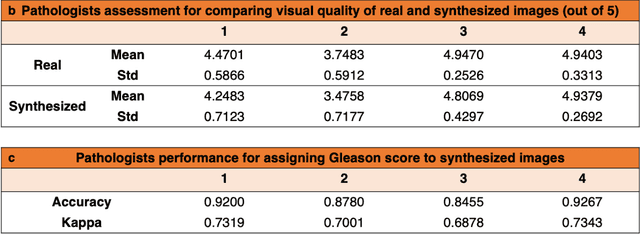
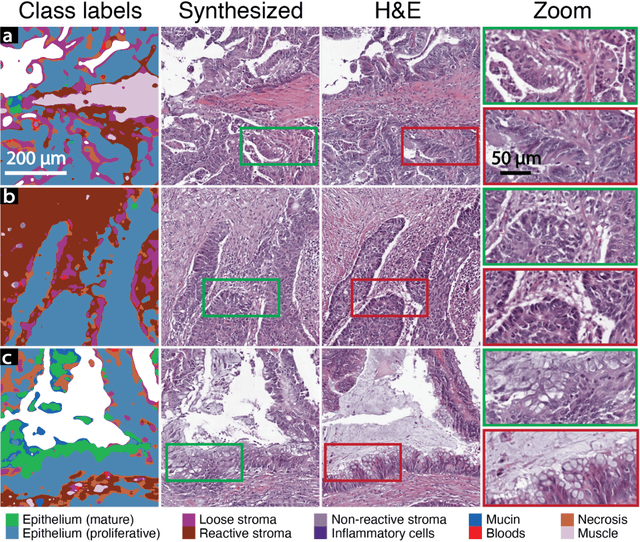
Abstract:An optical microscopic examination of thinly cut stained tissue on glass slides prepared from a FFPE tissue blocks is the gold standard for tissue diagnostics. In addition, the diagnostic abilities and expertise of any pathologist is dependent on their direct experience with common as well as rarer variant morphologies. Recently, deep learning approaches have been used to successfully show a high level of accuracy for such tasks. However, obtaining expert-level annotated images is an expensive and time-consuming task and artificially synthesized histological images can prove greatly beneficial. Here, we present an approach to not only generate histological images that reproduce the diagnostic morphologic features of common disease but also provide a user ability to generate new and rare morphologies. Our approach involves developing a generative adversarial network model that synthesizes pathology images constrained by class labels. We investigated the ability of this framework in synthesizing realistic prostate and colon tissue images and assessed the utility of these images in augmenting diagnostic ability of machine learning methods as well as their usability by a panel of experienced anatomic pathologists. Synthetic data generated by our framework performed similar to real data in training a deep learning model for diagnosis. Pathologists were not able to distinguish between real and synthetic images and showed a similar level of inter-observer agreement for prostate cancer grading. We extended the approach to significantly more complex images from colon biopsies and showed that the complex microenvironment in such tissues can also be reproduced. Finally, we present the ability for a user to generate deepfake histological images via a simple markup of sematic labels.
A generative adversarial approach to facilitate archival-quality histopathologic diagnoses from frozen tissue sections
Aug 24, 2021
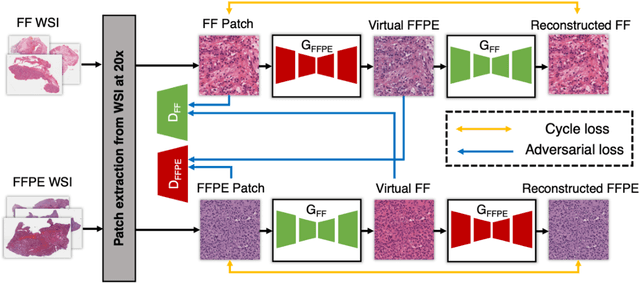

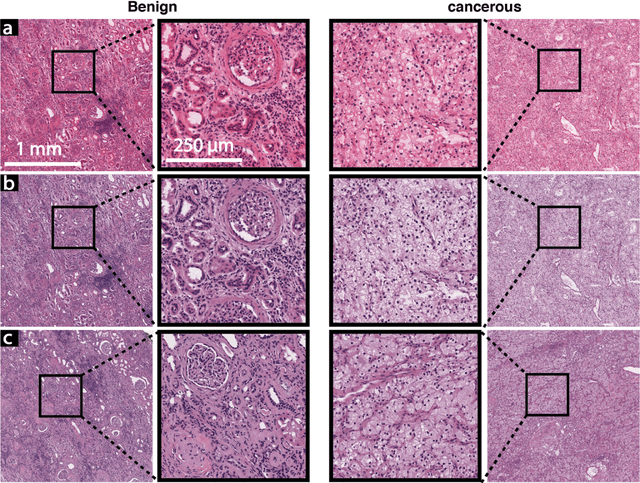
Abstract:In clinical diagnostics and research involving histopathology, formalin fixed paraffin embedded (FFPE) tissue is almost universally favored for its superb image quality. However, tissue processing time (more than 24 hours) can slow decision-making. In contrast, fresh frozen (FF) processing (less than 1 hour) can yield rapid information but diagnostic accuracy is suboptimal due to lack of clearing, morphologic deformation and more frequent artifacts. Here, we bridge this gap using artificial intelligence. We synthesize FFPE-like images ,virtual FFPE, from FF images using a generative adversarial network (GAN) from 98 paired kidney samples derived from 40 patients. Five board-certified pathologists evaluated the results in a blinded test. Image quality of the virtual FFPE data was assessed to be high and showed a close resemblance to real FFPE images. Clinical assessments of disease on the virtual FFPE images showed a higher inter-observer agreement compared to FF images. The nearly instantaneously generated virtual FFPE images can not only reduce time to information but can facilitate more precise diagnosis from routine FF images without extraneous costs and effort.
A deep learning framework for morphologic detail beyond the diffraction limit in infrared spectroscopic imaging
Dec 19, 2019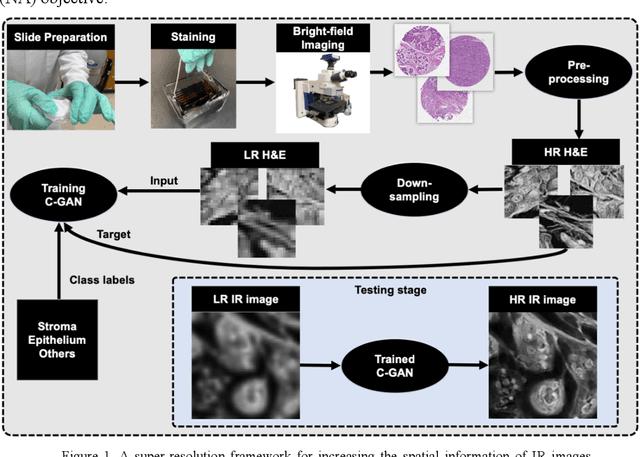
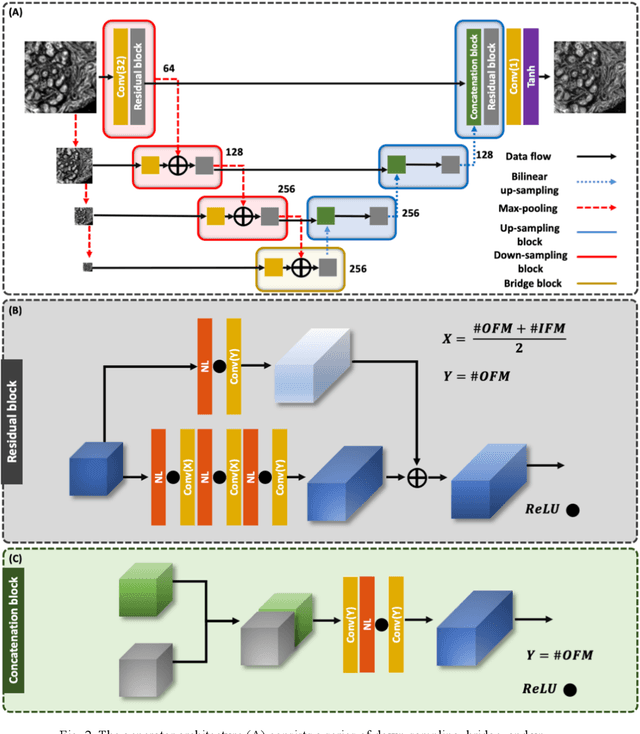
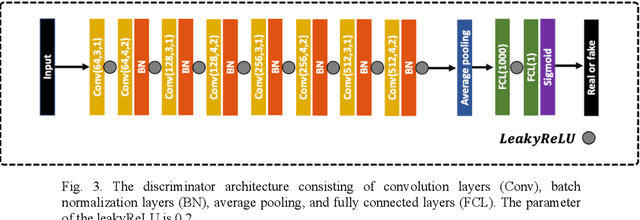
Abstract:Infrared (IR) microscopes measure spectral information that quantifies molecular content to assign the identity of biomedical cells but lack the spatial quality of optical microscopy to appreciate morphologic features. Here, we propose a method to utilize the semantic information of cellular identity from IR imaging with the morphologic detail of pathology images in a deep learning-based approach to image super-resolution. Using Generative Adversarial Networks (GANs), we enhance the spatial detail in IR imaging beyond the diffraction limit while retaining their spectral contrast. This technique can be rapidly integrated with modern IR microscopes to provide a framework useful for routine pathology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge