Kevin A. Schneider
Comparative Analysis of Quantum and Classical Support Vector Classifiers for Software Bug Prediction: An Exploratory Study
Jan 08, 2025Abstract:Purpose: Quantum computing promises to transform problem-solving across various domains with rapid and practical solutions. Within Software Evolution and Maintenance, Quantum Machine Learning (QML) remains mostly an underexplored domain, particularly in addressing challenges such as detecting buggy software commits from code repositories. Methods: In this study, we investigate the practical application of Quantum Support Vector Classifiers (QSVC) for detecting buggy software commits across 14 open-source software projects with diverse dataset sizes encompassing 30,924 data instances. We compare the QML algorithm PQSVC (Pegasos QSVC) and QSVC against the classical Support Vector Classifier (SVC). Our technique addresses large datasets in QSVC algorithms by dividing them into smaller subsets. We propose and evaluate an aggregation method to combine predictions from these models to detect the entire test dataset. We also introduce an incremental testing methodology to overcome the difficulties of quantum feature mapping during the testing approach. Results: The study shows the effectiveness of QSVC and PQSVC in detecting buggy software commits. The aggregation technique successfully combines predictions from smaller data subsets, enhancing the overall detection accuracy for the entire test dataset. The incremental testing methodology effectively manages the challenges associated with quantum feature mapping during the testing process. Conclusion: We contribute to the advancement of QML algorithms in defect prediction, unveiling the potential for further research in this domain. The specific scenario of the Short-Term Activity Frame (STAF) highlights the early detection of buggy software commits during the initial developmental phases of software systems, particularly when dataset sizes remain insufficient to train machine learning models.
Evaluating the Performance of a D-Wave Quantum Annealing System for Feature Subset Selection in Software Defect Prediction
Oct 21, 2024



Abstract:Predicting software defects early in the development process not only enhances the quality and reliability of the software but also decreases the cost of development. A wide range of machine learning techniques can be employed to create software defect prediction models, but the effectiveness and accuracy of these models are often influenced by the choice of appropriate feature subset. Since finding the optimal feature subset is computationally intensive, heuristic and metaheuristic approaches are commonly employed to identify near-optimal solutions within a reasonable time frame. Recently, the quantum computing paradigm quantum annealing (QA) has been deployed to find solutions to complex optimization problems. This opens up the possibility of addressing the feature subset selection problem with a QA machine. Although several strategies have been proposed for feature subset selection using a QA machine, little exploration has been done regarding the viability of a QA machine for feature subset selection in software defect prediction. This study investigates the potential of D-Wave QA system for this task, where we formulate a mutual information (MI)-based filter approach as an optimization problem and utilize a D-Wave Quantum Processing Unit (QPU) solver as a QA solver for feature subset selection. We evaluate the performance of this approach using multiple software defect datasets from the AEEM, JIRA, and NASA projects. We also utilize a D-Wave classical solver for comparative analysis. Our experimental results demonstrate that QA-based feature subset selection can enhance software defect prediction. Although the D-Wave QPU solver exhibits competitive prediction performance with the classical solver in software defect prediction, it significantly reduces the time required to identify the best feature subset compared to its classical counterpart.
Unveiling the potential of large language models in generating semantic and cross-language clones
Sep 12, 2023


Abstract:Semantic and Cross-language code clone generation may be useful for code reuse, code comprehension, refactoring and benchmarking. OpenAI's GPT model has potential in such clone generation as GPT is used for text generation. When developers copy/paste codes from Stack Overflow (SO) or within a system, there might be inconsistent changes leading to unexpected behaviours. Similarly, if someone possesses a code snippet in a particular programming language but seeks equivalent functionality in a different language, a semantic cross-language code clone generation approach could provide valuable assistance. In this study, using SemanticCloneBench as a vehicle, we evaluated how well the GPT-3 model could help generate semantic and cross-language clone variants for a given fragment.We have comprised a diverse set of code fragments and assessed GPT-3s performance in generating code variants.Through extensive experimentation and analysis, where 9 judges spent 158 hours to validate, we investigate the model's ability to produce accurate and semantically correct variants. Our findings shed light on GPT-3's strengths in code generation, offering insights into the potential applications and challenges of using advanced language models in software development. Our quantitative analysis yields compelling results. In the realm of semantic clones, GPT-3 attains an impressive accuracy of 62.14% and 0.55 BLEU score, achieved through few-shot prompt engineering. Furthermore, the model shines in transcending linguistic confines, boasting an exceptional 91.25% accuracy in generating cross-language clones
Automatic Polyp Segmentation Using Convolutional Neural Networks
Apr 22, 2020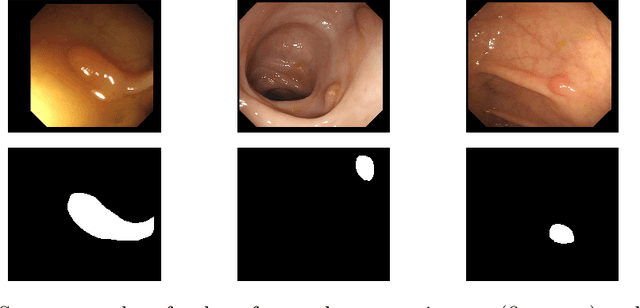



Abstract:Colorectal cancer is the third most common cancer-related death after lung cancer and breast cancer worldwide. The risk of developing colorectal cancer could be reduced by early diagnosis of polyps during a colonoscopy. Computer-aided diagnosis systems have the potential to be applied for polyp screening and reduce the number of missing polyps. In this paper, we compare the performance of different deep learning architectures as feature extractors, i.e. ResNet, DenseNet, InceptionV3, InceptionResNetV2 and SE-ResNeXt in the encoder part of a U-Net architecture. We validated the performance of presented ensemble models on the CVC-Clinic (GIANA 2018) dataset. The DenseNet169 feature extractor combined with U-Net architecture outperformed the other counterparts and achieved an accuracy of 99.15\%, Dice similarity coefficient of 90.87%, and Jaccard index of 83.82%.
Automatic Detection of Coronavirus Disease (COVID-19) in X-ray and CT Images: A Machine Learning-Based Approach
Apr 22, 2020
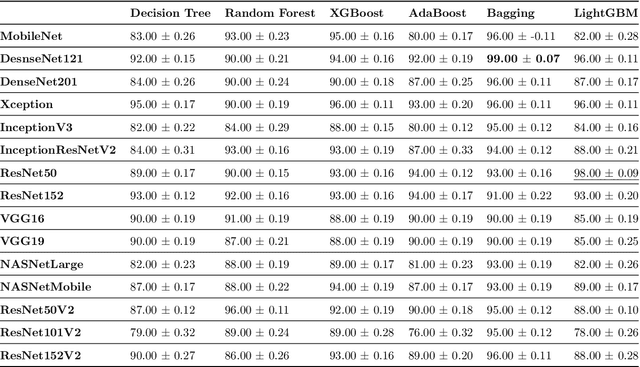

Abstract:The newly identified Coronavirus pneumonia, subsequently termed COVID-19, is highly transmittable and pathogenic with no clinically approved antiviral drug or vaccine available for treatment. The most common symptoms of COVID-19 are dry cough, sore throat, and fever. Symptoms can progress to a severe form of pneumonia with critical complications, including septic shock, pulmonary edema, acute respiratory distress syndrome and multi-organ failure. While medical imaging is not currently recommended in Canada for primary diagnosis of COVID-19, computer-aided diagnosis systems could assist in the early detection of COVID-19 abnormalities and help to monitor the progression of the disease, potentially reduce mortality rates. In this study, we compare popular deep learning-based feature extraction frameworks for automatic COVID-19 classification. To obtain the most accurate feature, which is an essential component of learning, MobileNet, DenseNet, Xception, ResNet, InceptionV3, InceptionResNetV2, VGGNet, NASNet were chosen amongst a pool of deep convolutional neural networks. The extracted features were then fed into several machine learning classifiers to classify subjects as either a case of COVID-19 or a control. This approach avoided task-specific data pre-processing methods to support a better generalization ability for unseen data. The performance of the proposed method was validated on a publicly available COVID-19 dataset of chest X-ray and CT images. The DenseNet121 feature extractor with Bagging tree classifier achieved the best performance with 99% classification accuracy. The second-best learner was a hybrid of the a ResNet50 feature extractor trained by LightGBM with an accuracy of 98%.
Classification of Histopathological Biopsy Images Using Ensemble of Deep Learning Networks
Sep 26, 2019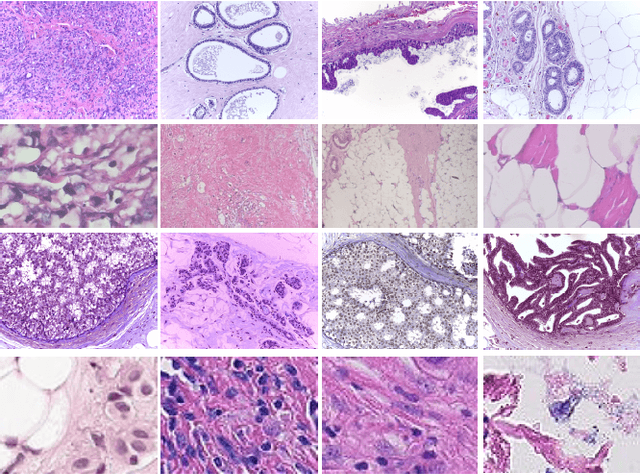
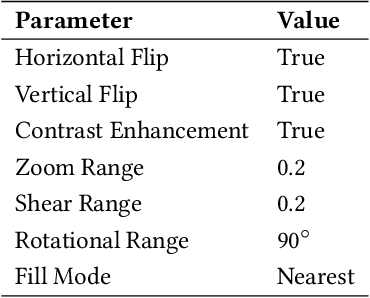


Abstract:Breast cancer is one of the leading causes of death across the world in women. Early diagnosis of this type of cancer is critical for treatment and patient care. Computer-aided detection (CAD) systems using convolutional neural networks (CNN) could assist in the classification of abnormalities. In this study, we proposed an ensemble deep learning-based approach for automatic binary classification of breast histology images. The proposed ensemble model adapts three pre-trained CNNs, namely VGG19, MobileNet, and DenseNet. The ensemble model is used for the feature representation and extraction steps. The extracted features are then fed into a multi-layer perceptron classifier to carry out the classification task. Various pre-processing and CNN tuning techniques such as stain-normalization, data augmentation, hyperparameter tuning, and fine-tuning are used to train the model. The proposed method is validated on four publicly available benchmark datasets, i.e., ICIAR, BreakHis, PatchCamelyon, and Bioimaging. The proposed multi-model ensemble method obtains better predictions than single classifiers and machine learning algorithms with accuracies of 98.13%, 95.00%, 94.64% and 83.10% for BreakHis, ICIAR, PatchCamelyon and Bioimaging datasets, respectively.
A Hybrid Deep Learning Architecture for Leukemic B-lymphoblast Classification
Sep 26, 2019



Abstract:Automatic detection of leukemic B-lymphoblast cancer in microscopic images is very challenging due to the complicated nature of histopathological structures. To tackle this issue, an automatic and robust diagnostic system is required for early detection and treatment. In this paper, an automated deep learning-based method is proposed to distinguish between immature leukemic blasts and normal cells. The proposed deep learning based hybrid method, which is enriched by different data augmentation techniques, is able to extract high-level features from input images. Results demonstrate that the proposed model yields better prediction than individual models for Leukemic B-lymphoblast classification with 96.17% overall accuracy, 95.17% sensitivity and 98.58% specificity. Fusing the features extracted from intermediate layers, our approach has the potential to improve the overall classification performance.
Breast Cancer Diagnosis with Transfer Learning and Global Pooling
Sep 26, 2019
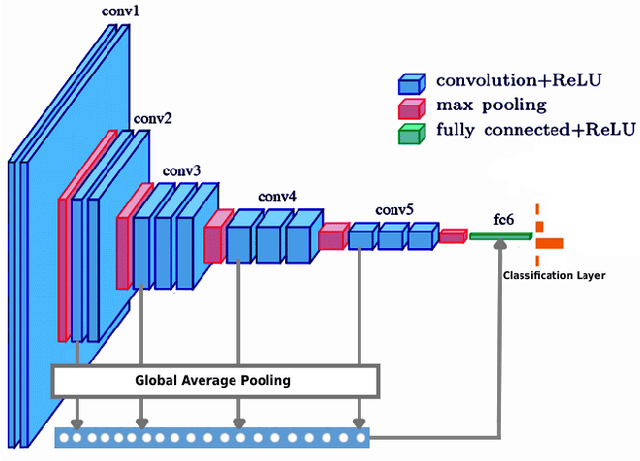
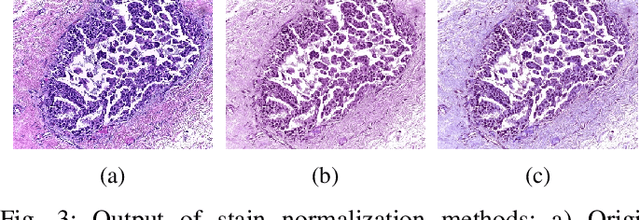
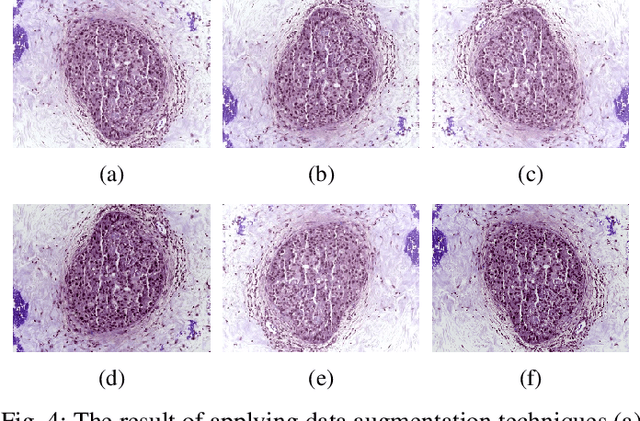
Abstract:Breast cancer is one of the most common causes of cancer-related death in women worldwide. Early and accurate diagnosis of breast cancer may significantly increase the survival rate of patients. In this study, we aim to develop a fully automatic, deep learning-based, method using descriptor features extracted by Deep Convolutional Neural Network (DCNN) models and pooling operation for the classification of hematoxylin and eosin stain (H&E) histological breast cancer images provided as a part of the International Conference on Image Analysis and Recognition (ICIAR) 2018 Grand Challenge on BreAst Cancer Histology (BACH) Images. Different data augmentation methods are applied to optimize the DCNN performance. We also investigated the efficacy of different stain normalization methods as a pre-processing step. The proposed network architecture using a pre-trained Xception model yields 92.50% average classification accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge