Keren Peri-Hanania
Detecting Bone Lesions in X-Ray Under Diverse Acquisition Conditions
Dec 15, 2022Abstract:The diagnosis of primary bone tumors is challenging, as the initial complaints are often non-specific. Early detection of bone cancer is crucial for a favorable prognosis. Incidentally, lesions may be found on radiographs obtained for other reasons. However, these early indications are often missed. In this work, we propose an automatic algorithm to detect bone lesions in conventional radiographs to facilitate early diagnosis. Detecting lesions in such radiographs is challenging: first, the prevalence of bone cancer is very low; any method must show high precision to avoid a prohibitive number of false alarms. Second, radiographs taken in health maintenance organizations (HMOs) or emergency departments (EDs) suffer from inherent diversity due to different X-ray machines, technicians and imaging protocols. This diversity poses a major challenge to any automatic analysis method. We propose to train an off-the-shelf object detection algorithm to detect lesions in radiographs. The novelty of our approach stems from a dedicated preprocessing stage that directly addresses the diversity of the data. The preprocessing consists of self-supervised region-of-interest detection using vision transformer (ViT), and a foreground-based histogram equalization for contrast enhancement to relevant regions only. We evaluate our method via a retrospective study that analyzes bone tumors on radiographs acquired from January 2003 to December 2018 under diverse acquisition protocols. Our method obtains 82.43% sensitivity at 1.5% false-positive rate and surpasses existing preprocessing methods. For lesion detection, our method achieves 82.5% accuracy and an IoU of 0.69. The proposed preprocessing method enables to effectively cope with the inherent diversity of radiographs acquired in HMOs and EDs.
Learned super resolution ultrasound for improved breast lesion characterization
Jul 12, 2021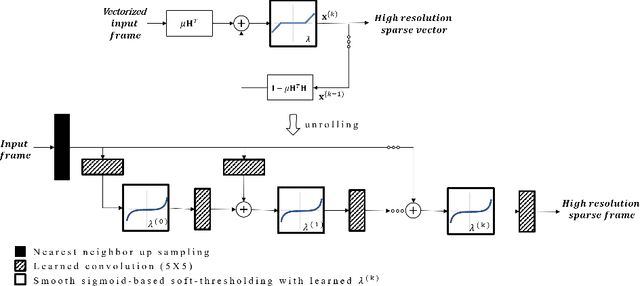
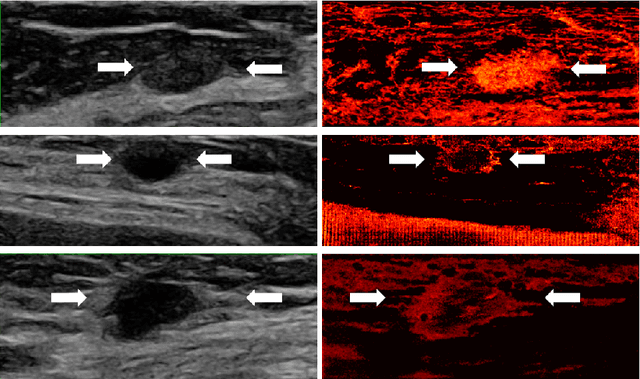
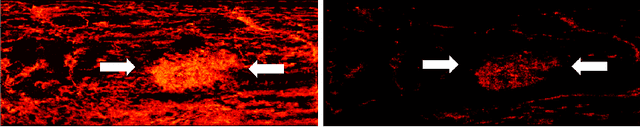
Abstract:Breast cancer is the most common malignancy in women. Mammographic findings such as microcalcifications and masses, as well as morphologic features of masses in sonographic scans, are the main diagnostic targets for tumor detection. However, improved specificity of these imaging modalities is required. A leading alternative target is neoangiogenesis. When pathological, it contributes to the development of numerous types of tumors, and the formation of metastases. Hence, demonstrating neoangiogenesis by visualization of the microvasculature may be of great importance. Super resolution ultrasound localization microscopy enables imaging of the microvasculature at the capillary level. Yet, challenges such as long reconstruction time, dependency on prior knowledge of the system Point Spread Function (PSF), and separability of the Ultrasound Contrast Agents (UCAs), need to be addressed for translation of super-resolution US into the clinic. In this work we use a deep neural network architecture that makes effective use of signal structure to address these challenges. We present in vivo human results of three different breast lesions acquired with a clinical US scanner. By leveraging our trained network, the microvasculature structure is recovered in a short time, without prior PSF knowledge, and without requiring separability of the UCAs. Each of the recoveries exhibits a different structure that corresponds with the known histological structure. This study demonstrates the feasibility of in vivo human super resolution, based on a clinical scanner, to increase US specificity for different breast lesions and promotes the use of US in the diagnosis of breast pathologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge