Kateryna Melnyk
Per Subject Complexity in Eye Movement Prediction
Dec 31, 2024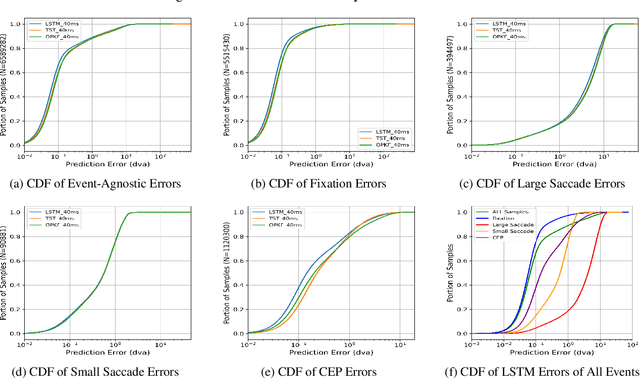

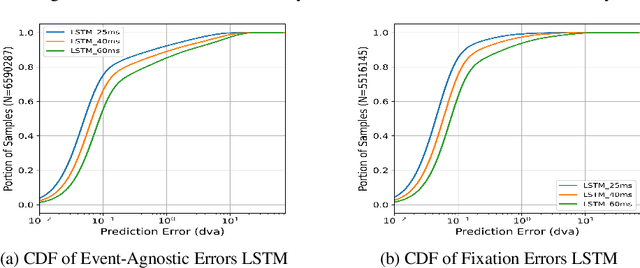

Abstract:Eye movement prediction is a promising area of research to compensate for the latency introduced by eye-tracking systems in virtual reality devices. In this study, we comprehensively analyze the complexity of the eye movement prediction task associated with subjects. We use three fundamentally different models within the analysis: the lightweight Long Short-Term Memory network (LSTM), the transformer-based network for multivariate time series representation learning (TST), and the Oculomotor Plant Mathematical Model wrapped in the Kalman Filter framework (OPKF). Each solution is assessed following a sample-to-event evaluation strategy and employing the new event-to-subject metrics. Our results show that the different models maintained similar prediction performance trends pertaining to subjects. We refer to these outcomes as per-subject complexity since some subjects' data pose a more significant challenge for models. Along with the detailed correlation analysis, this report investigates the source of the per-subject complexity and discusses potential solutions to overcome it.
Understanding microbiome dynamics via interpretable graph representation learning
Mar 02, 2022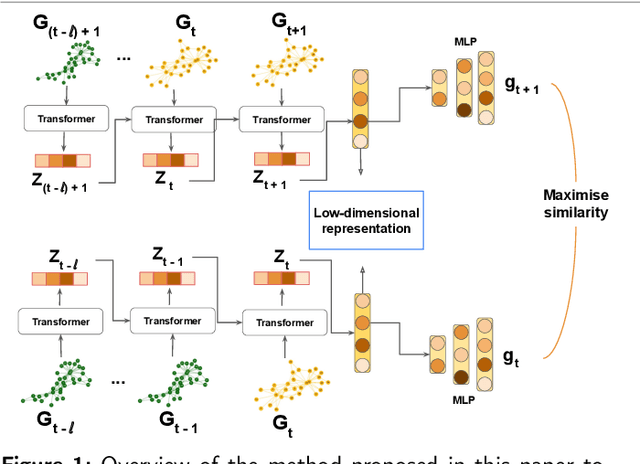
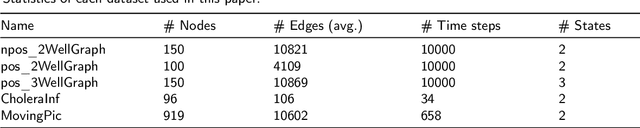
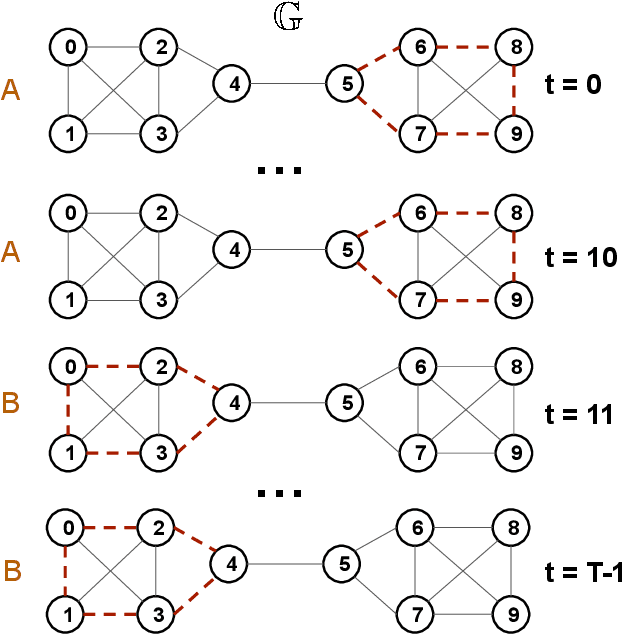
Abstract:Large-scale perturbations in the microbiome constitution are strongly correlated, whether as a driver or a consequence, with the health and functioning of human physiology. However, understanding the difference in the microbiome profiles of healthy and ill individuals can be complicated due to the large number of complex interactions among microbes. We propose to model these interactions as a time-evolving graph whose nodes are microbes and edges are interactions among them. Motivated by the need to analyse such complex interactions, we develop a method that learns a low-dimensional representation of the time-evolving graph and maintains the dynamics occurring in the high-dimensional space. Through our experiments, we show that we can extract graph features such as clusters of nodes or edges that have the highest impact on the model to learn the low-dimensional representation. This information can be crucial to identify microbes and interactions among them that are strongly correlated with clinical diseases. We conduct our experiments on both synthetic and real-world microbiome datasets.
GraphKKE: Graph Kernel Koopman Embedding for Human Microbiome Analysis
Sep 07, 2020
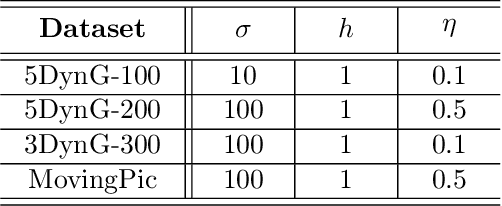
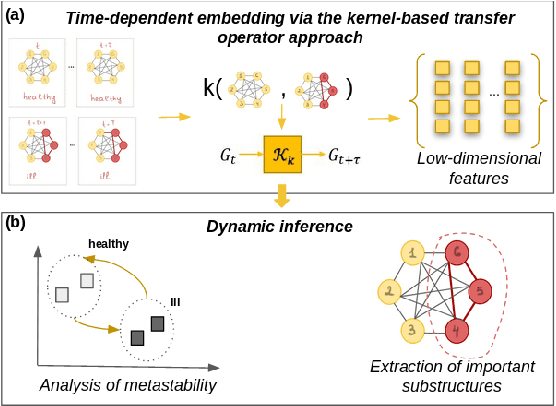

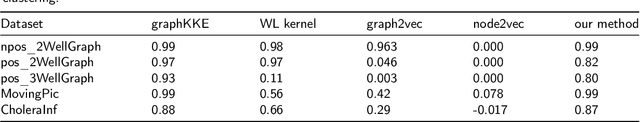
Abstract:More and more diseases have been found to be strongly correlated with disturbances in the microbiome constitution, e.g., obesity, diabetes, or some cancer types. Thanks to modern high-throughput omics technologies, it becomes possible to directly analyze human microbiome and its influence on the health status. Microbial communities are monitored over long periods of time and the associations between their members are explored. These relationships can be described by a time-evolving graph. In order to understand responses of the microbial community members to a distinct range of perturbations such as antibiotics exposure or diseases and general dynamical properties, the time-evolving graph of the human microbial communities has to be analyzed. This becomes especially challenging due to dozens of complex interactions among microbes and metastable dynamics. The key to solving this problem is the representation of the time-evolving graphs as fixed-length feature vectors preserving the original dynamics. We propose a method for learning the embedding of the time-evolving graph that is based on the spectral analysis of transfer operators and graph kernels. We demonstrate that our method can capture temporary changes in the time-evolving graph on both created synthetic data and real-world data. Our experiments demonstrate the efficacy of the method. Furthermore, we show that our method can be applied to human microbiome data to study dynamic processes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge