Tim O. F. Conrad
Combining LLMs and Knowledge Graphs to Reduce Hallucinations in Question Answering
Sep 06, 2024Abstract:Advancements in natural language processing have revolutionized the way we can interact with digital information systems, such as databases, making them more accessible. However, challenges persist, especially when accuracy is critical, as in the biomedical domain. A key issue is the hallucination problem, where models generate information unsupported by the underlying data, potentially leading to dangerous misinformation. This paper presents a novel approach designed to bridge this gap by combining Large Language Models (LLM) and Knowledge Graphs (KG) to improve the accuracy and reliability of question-answering systems, on the example of a biomedical KG. Built on the LangChain framework, our method incorporates a query checker that ensures the syntactical and semantic validity of LLM-generated queries, which are then used to extract information from a Knowledge Graph, substantially reducing errors like hallucinations. We evaluated the overall performance using a new benchmark dataset of 50 biomedical questions, testing several LLMs, including GPT-4 Turbo and llama3:70b. Our results indicate that while GPT-4 Turbo outperforms other models in generating accurate queries, open-source models like llama3:70b show promise with appropriate prompt engineering. To make this approach accessible, a user-friendly web-based interface has been developed, allowing users to input natural language queries, view generated and corrected Cypher queries, and verify the resulting paths for accuracy. Overall, this hybrid approach effectively addresses common issues such as data gaps and hallucinations, offering a reliable and intuitive solution for question answering systems. The source code for generating the results of this paper and for the user-interface can be found in our Git repository: https://git.zib.de/lpusch/cyphergenkg-gui
Forget Embedding Layers: Representation Learning for Cold-start in Recommender Systems
Oct 30, 2022Abstract:Recommender systems suffer from the cold-start problem whenever a new user joins the platform or a new item is added to the catalog. To address item cold-start, we propose to replace the embedding layer in sequential recommenders with a dynamic storage that has no learnable weights and can keep an arbitrary number of representations. In this paper, we present FELRec, a large embedding network that refines the existing representations of users and items in a recursive manner, as new information becomes available. In contrast to similar approaches, our model represents new users and items without side information or time-consuming fine-tuning. During item cold-start, our method outperforms similar method by 29.50%-47.45%. Further, our proposed model generalizes well to previously unseen datasets. The source code is publicly available at github.com/kweimann/FELRec.
Understanding microbiome dynamics via interpretable graph representation learning
Mar 02, 2022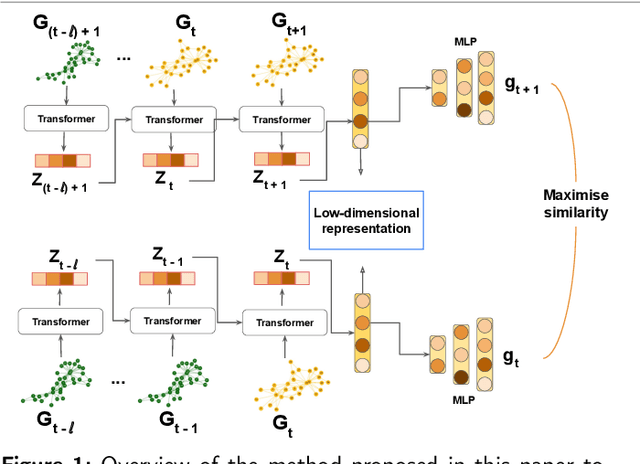
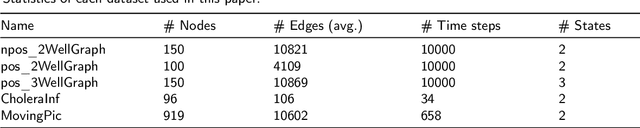
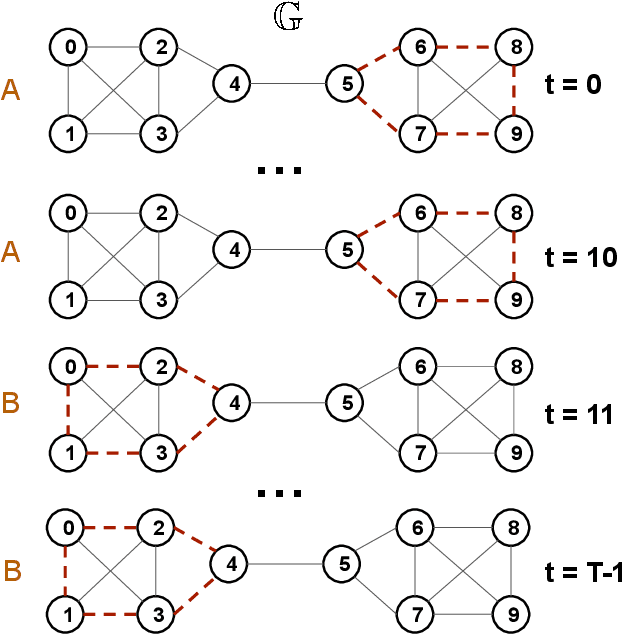
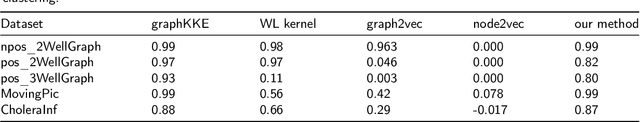
Abstract:Large-scale perturbations in the microbiome constitution are strongly correlated, whether as a driver or a consequence, with the health and functioning of human physiology. However, understanding the difference in the microbiome profiles of healthy and ill individuals can be complicated due to the large number of complex interactions among microbes. We propose to model these interactions as a time-evolving graph whose nodes are microbes and edges are interactions among them. Motivated by the need to analyse such complex interactions, we develop a method that learns a low-dimensional representation of the time-evolving graph and maintains the dynamics occurring in the high-dimensional space. Through our experiments, we show that we can extract graph features such as clusters of nodes or edges that have the highest impact on the model to learn the low-dimensional representation. This information can be crucial to identify microbes and interactions among them that are strongly correlated with clinical diseases. We conduct our experiments on both synthetic and real-world microbiome datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge