Kuba Weimann
Forget Embedding Layers: Representation Learning for Cold-start in Recommender Systems
Oct 30, 2022Abstract:Recommender systems suffer from the cold-start problem whenever a new user joins the platform or a new item is added to the catalog. To address item cold-start, we propose to replace the embedding layer in sequential recommenders with a dynamic storage that has no learnable weights and can keep an arbitrary number of representations. In this paper, we present FELRec, a large embedding network that refines the existing representations of users and items in a recursive manner, as new information becomes available. In contrast to similar approaches, our model represents new users and items without side information or time-consuming fine-tuning. During item cold-start, our method outperforms similar method by 29.50%-47.45%. Further, our proposed model generalizes well to previously unseen datasets. The source code is publicly available at github.com/kweimann/FELRec.
Understanding microbiome dynamics via interpretable graph representation learning
Mar 02, 2022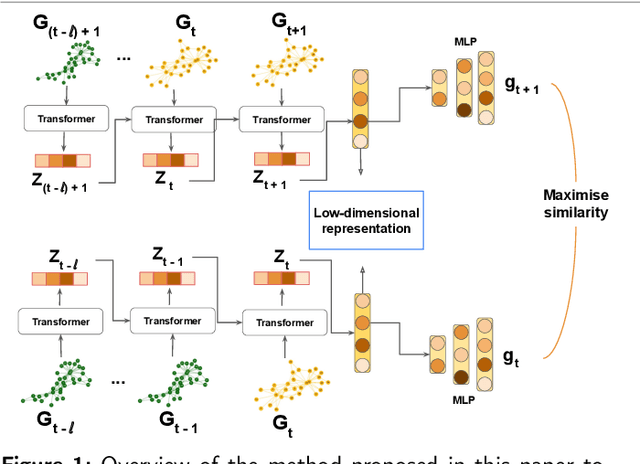
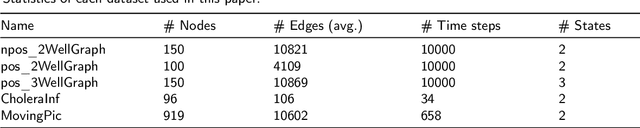
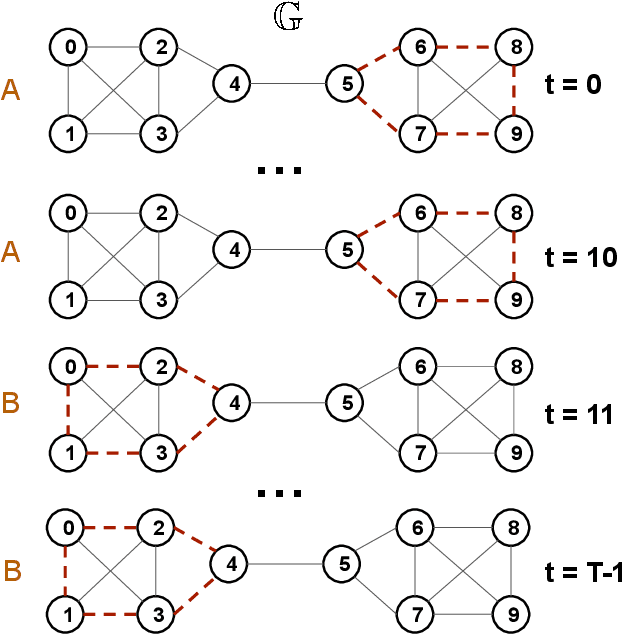
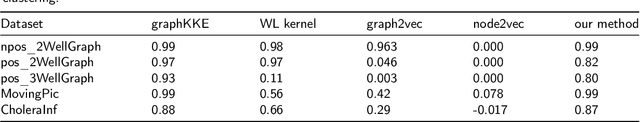
Abstract:Large-scale perturbations in the microbiome constitution are strongly correlated, whether as a driver or a consequence, with the health and functioning of human physiology. However, understanding the difference in the microbiome profiles of healthy and ill individuals can be complicated due to the large number of complex interactions among microbes. We propose to model these interactions as a time-evolving graph whose nodes are microbes and edges are interactions among them. Motivated by the need to analyse such complex interactions, we develop a method that learns a low-dimensional representation of the time-evolving graph and maintains the dynamics occurring in the high-dimensional space. Through our experiments, we show that we can extract graph features such as clusters of nodes or edges that have the highest impact on the model to learn the low-dimensional representation. This information can be crucial to identify microbes and interactions among them that are strongly correlated with clinical diseases. We conduct our experiments on both synthetic and real-world microbiome datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge