Karen Hodgson
Contextualised concept embedding for efficiently adapting natural language processing models for phenotype identification
Mar 13, 2019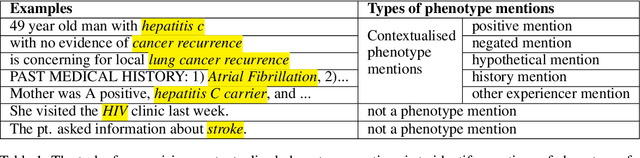
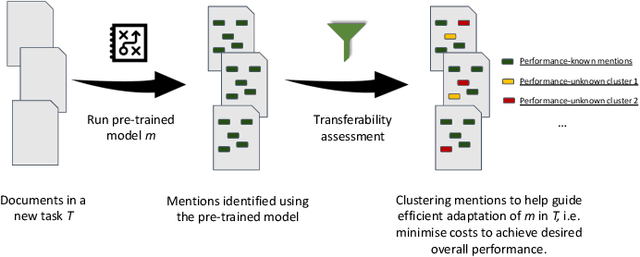
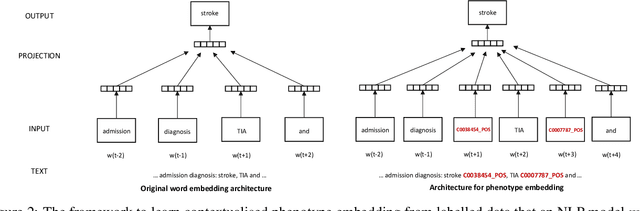
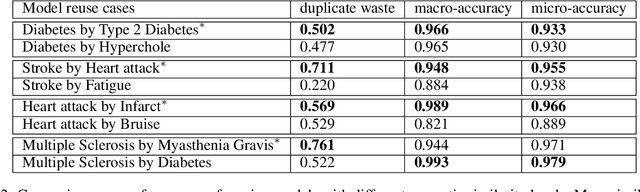
Abstract:Many efforts have been put to use automated approaches, such as natural language processing (NLP), to mine or extract data from free-text medical records to picture comprehensive patient profiles for delivering better health-care. Reusing NLP models in new settings, however, remains cumbersome - requiring validation and/or retraining on new data iteratively to achieve convergent results. In this paper, we formally define and analyse the NLP model adaptation problem, particularly in phenotype identification tasks, and identify two types of common unnecessary or wasted efforts: duplicate waste and imbalance waste. A distributed representation approach is proposed to represent familiar language patterns for an NLP model by learning phenotype embeddings from its training data. Computations on these language patterns are then introduced to help avoid or reduce unnecessary efforts by combining both geometric and semantic similarities. To evaluate the approach, we cross validate NLP models developed for six physical morbidity studies (23 phenotypes; 17 million documents) on anonymised medical records of South London Maudsley NHS Trust, United Kingdom. Two metrics are introduced to quantify the reductions for both duplicate and imbalance wastes. We conducted various experiments on reusing NLP models in four phenotype identification tasks. Our approach can choose a best model for a given new task, which can identify up to 76% mentions needing no validation & model retraining, meanwhile, having very good performances (93-97% accuracy). It can also provide guidance for validating and retraining the model for novel language patterns in new tasks, which can help save around 80% of the efforts required in blind model-adaptation approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge