Junchen Yang
Multi-modal Differentiable Unsupervised Feature Selection
Mar 16, 2023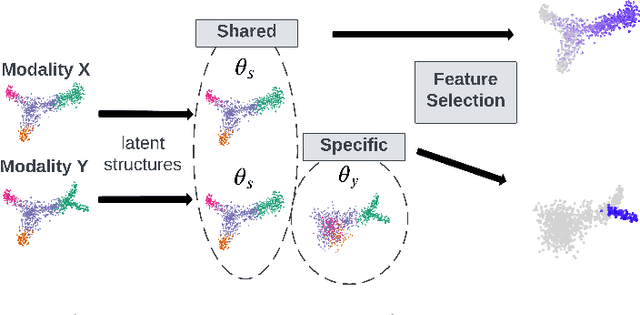
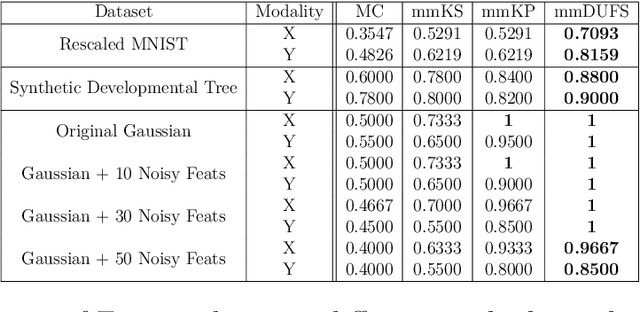
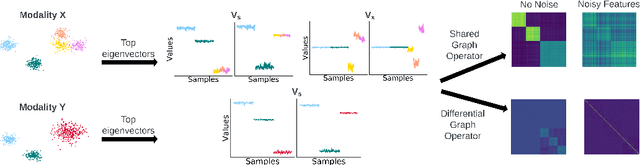
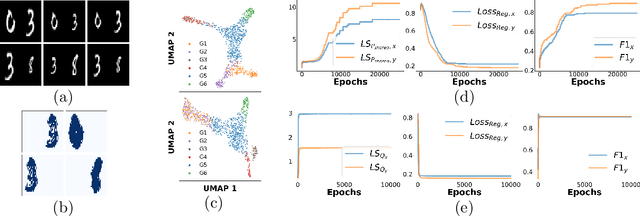
Abstract:Multi-modal high throughput biological data presents a great scientific opportunity and a significant computational challenge. In multi-modal measurements, every sample is observed simultaneously by two or more sets of sensors. In such settings, many observed variables in both modalities are often nuisance and do not carry information about the phenomenon of interest. Here, we propose a multi-modal unsupervised feature selection framework: identifying informative variables based on coupled high-dimensional measurements. Our method is designed to identify features associated with two types of latent low-dimensional structures: (i) shared structures that govern the observations in both modalities and (ii) differential structures that appear in only one modality. To that end, we propose two Laplacian-based scoring operators. We incorporate the scores with differentiable gates that mask nuisance features and enhance the accuracy of the structure captured by the graph Laplacian. The performance of the new scheme is illustrated using synthetic and real datasets, including an extended biological application to single-cell multi-omics.
Locally Sparse Networks for Interpretable Predictions
Jun 11, 2021



Abstract:Despite the enormous success of neural networks, they are still hard to interpret and often overfit when applied to low-sample-size (LSS) datasets. To tackle these obstacles, we propose a framework for training locally sparse neural networks where the local sparsity is learned via a sample-specific gating mechanism that identifies the subset of most relevant features for each measurement. The sample-specific sparsity is predicted via a \textit{gating} network, which is trained in tandem with the \textit{prediction} network. By learning these subsets and weights of a prediction model, we obtain an interpretable neural network that can handle LSS data and can remove nuisance variables, which are irrelevant for the supervised learning task. Using both synthetic and real-world datasets, we demonstrate that our method outperforms state-of-the-art models when predicting the target function with far fewer features per instance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge