Juliane Fluck
NFDI4Health workflow and service for synthetic data generation, assessment and risk management
Aug 08, 2024



Abstract:Individual health data is crucial for scientific advancements, particularly in developing Artificial Intelligence (AI); however, sharing real patient information is often restricted due to privacy concerns. A promising solution to this challenge is synthetic data generation. This technique creates entirely new datasets that mimic the statistical properties of real data, while preserving confidential patient information. In this paper, we present the workflow and different services developed in the context of Germany's National Data Infrastructure project NFDI4Health. First, two state-of-the-art AI tools (namely, VAMBN and MultiNODEs) for generating synthetic health data are outlined. Further, we introduce SYNDAT (a public web-based tool) which allows users to visualize and assess the quality and risk of synthetic data provided by desired generative models. Additionally, the utility of the proposed methods and the web-based tool is showcased using data from Alzheimer's Disease Neuroimaging Initiative (ADNI) and the Center for Cancer Registry Data of the Robert Koch Institute (RKI).
Synthetic data generation for a longitudinal cohort study -- Evaluation, method extension and reproduction of published data analysis results
May 12, 2023Abstract:Access to individual-level health data is essential for gaining new insights and advancing science. In particular, modern methods based on artificial intelligence rely on the availability of and access to large datasets. In the health sector, access to individual-level data is often challenging due to privacy concerns. A promising alternative is the generation of fully synthetic data, i.e. data generated through a randomised process that have similar statistical properties as the original data, but do not have a one-to-one correspondence with the original individual-level records. In this study, we use a state-of-the-art synthetic data generation method and perform in-depth quality analyses of the generated data for a specific use case in the field of nutrition. We demonstrate the need for careful analyses of synthetic data that go beyond descriptive statistics and provide valuable insights into how to realise the full potential of synthetic datasets. By extending the methods, but also by thoroughly analysing the effects of sampling from a trained model, we are able to largely reproduce significant real-world analysis results in the chosen use case.
BERT WEAVER: Using WEight AVERaging to Enable Lifelong Learning for Transformer-based Models
Feb 21, 2022
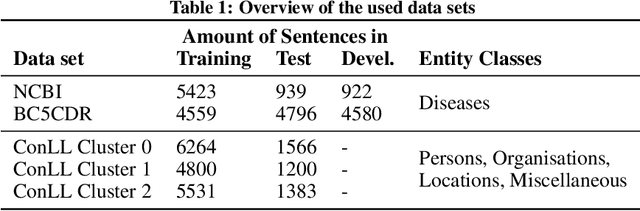

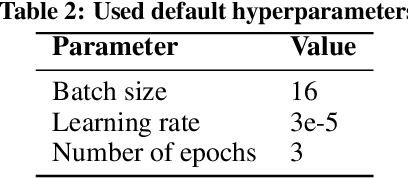
Abstract:Recent developments in transfer learning have boosted the advancements in natural language processing tasks. The performance is, however, dependent on high-quality, manually annotated training data. Especially in the biomedical domain, it has been shown that one training corpus is not enough to learn generic models that are able to efficiently predict on new data. Therefore, state-of-the-art models need the ability of lifelong learning in order to improve performance as soon as new data are available - without the need of retraining the whole model from scratch. We present WEAVER, a simple, yet efficient post-processing method that infuses old knowledge into the new model, thereby reducing catastrophic forgetting. We show that applying WEAVER in a sequential manner results in similar word embedding distributions as doing a combined training on all data at once, while being computationally more efficient. Because there is no need of data sharing, the presented method is also easily applicable to federated learning settings and can for example be beneficial for the mining of electronic health records from different clinics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge