Holger Fröhlich
Improving Representation Learning of Complex Critical Care Data with ICU-BERT
Feb 26, 2025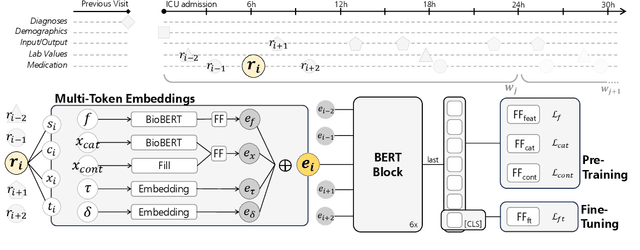
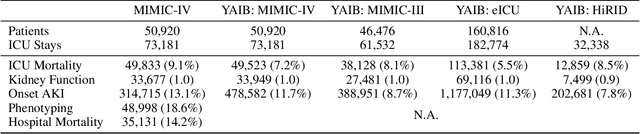
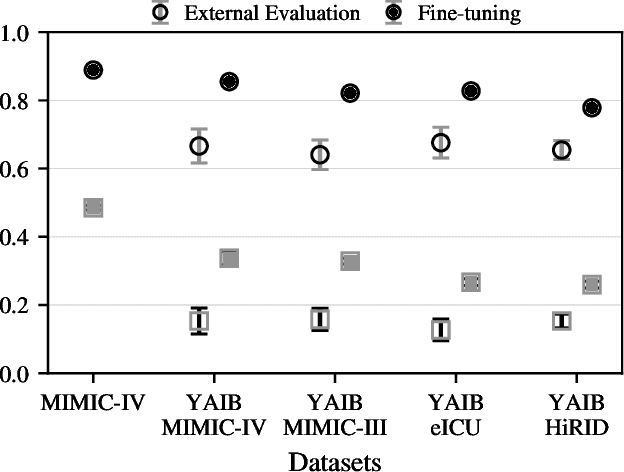
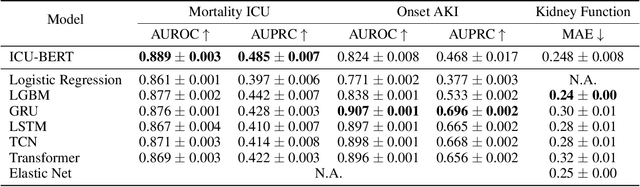
Abstract:The multivariate, asynchronous nature of real-world clinical data, such as that generated in Intensive Care Units (ICUs), challenges traditional AI-based decision-support systems. These often assume data regularity and feature independence and frequently rely on limited data scopes and manual feature engineering. The potential of generative AI technologies has not yet been fully exploited to analyze clinical data. We introduce ICU-BERT, a transformer-based model pre-trained on the MIMIC-IV database using a multi-task scheme to learn robust representations of complex ICU data with minimal preprocessing. ICU-BERT employs a multi-token input strategy, incorporating dense embeddings from a biomedical Large Language Model to learn a generalizable representation of complex and multivariate ICU data. With an initial evaluation of five tasks and four additional ICU datasets, ICU-BERT results indicate that ICU-BERT either compares to or surpasses current performance benchmarks by leveraging fine-tuning. By integrating structured and unstructured data, ICU-BERT advances the use of foundational models in medical informatics, offering an adaptable solution for clinical decision support across diverse applications.
NFDI4Health workflow and service for synthetic data generation, assessment and risk management
Aug 08, 2024



Abstract:Individual health data is crucial for scientific advancements, particularly in developing Artificial Intelligence (AI); however, sharing real patient information is often restricted due to privacy concerns. A promising solution to this challenge is synthetic data generation. This technique creates entirely new datasets that mimic the statistical properties of real data, while preserving confidential patient information. In this paper, we present the workflow and different services developed in the context of Germany's National Data Infrastructure project NFDI4Health. First, two state-of-the-art AI tools (namely, VAMBN and MultiNODEs) for generating synthetic health data are outlined. Further, we introduce SYNDAT (a public web-based tool) which allows users to visualize and assess the quality and risk of synthetic data provided by desired generative models. Additionally, the utility of the proposed methods and the web-based tool is showcased using data from Alzheimer's Disease Neuroimaging Initiative (ADNI) and the Center for Cancer Registry Data of the Robert Koch Institute (RKI).
Towards early diagnosis of Alzheimer's disease: Advances in immune-related blood biomarkers and computational modeling approaches
Dec 06, 2023Abstract:Alzheimer's disease has an increasing prevalence in the population world-wide, yet current diagnostic methods based on recommended biomarkers are only available in specialized clinics. Due to these circumstances, Alzheimer's disease is usually diagnosed late, which contrasts with the currently available treatment options that are only effective for patients at an early stage. Blood-based biomarkers could fill in the gap of easily accessible and low-cost methods for early diagnosis of the disease. In particular, immune-based blood-biomarkers might be a promising option, given the recently discovered cross-talk of immune cells of the central nervous system with those in the peripheral immune system. With the help of machine learning algorithms and mechanistic modeling approaches, such as agent-based modeling, an in-depth analysis of the simulation of cell dynamics is possible as well as of high-dimensional omics resources indicative of pathway signaling changes. Here, we give a background on advances in research on brain-immune system cross-talk in Alzheimer's disease and review recent machine learning and mechanistic modeling approaches which leverage modern omics technologies for blood-based immune system-related biomarker discovery.
Synthetic data generation for a longitudinal cohort study -- Evaluation, method extension and reproduction of published data analysis results
May 12, 2023Abstract:Access to individual-level health data is essential for gaining new insights and advancing science. In particular, modern methods based on artificial intelligence rely on the availability of and access to large datasets. In the health sector, access to individual-level data is often challenging due to privacy concerns. A promising alternative is the generation of fully synthetic data, i.e. data generated through a randomised process that have similar statistical properties as the original data, but do not have a one-to-one correspondence with the original individual-level records. In this study, we use a state-of-the-art synthetic data generation method and perform in-depth quality analyses of the generated data for a specific use case in the field of nutrition. We demonstrate the need for careful analyses of synthetic data that go beyond descriptive statistics and provide valuable insights into how to realise the full potential of synthetic datasets. By extending the methods, but also by thoroughly analysing the effects of sampling from a trained model, we are able to largely reproduce significant real-world analysis results in the chosen use case.
Integrating Prior Knowledge Into Prognostic Biomarker Discovery based on Network Structure
May 27, 2013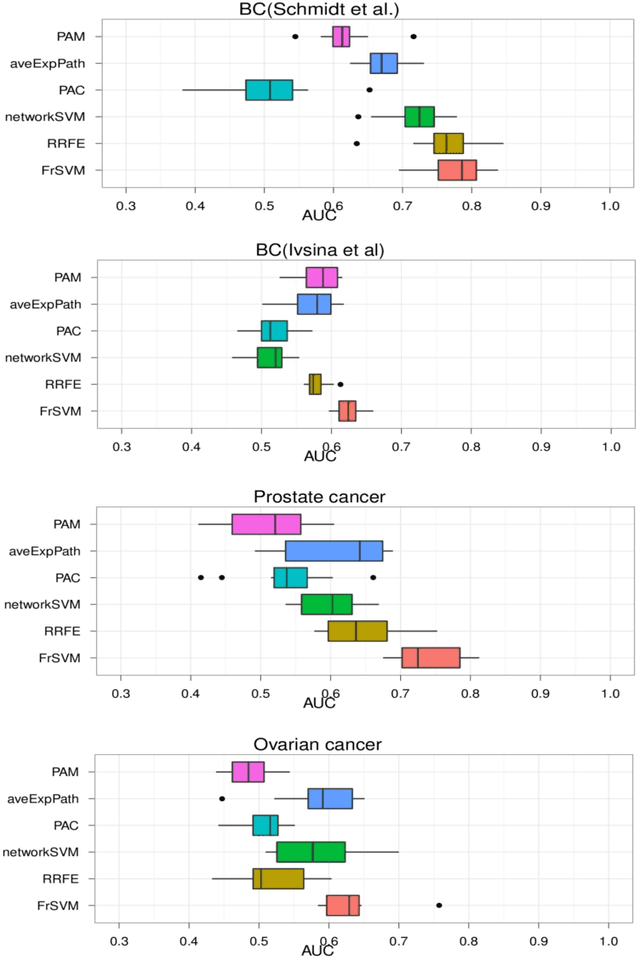
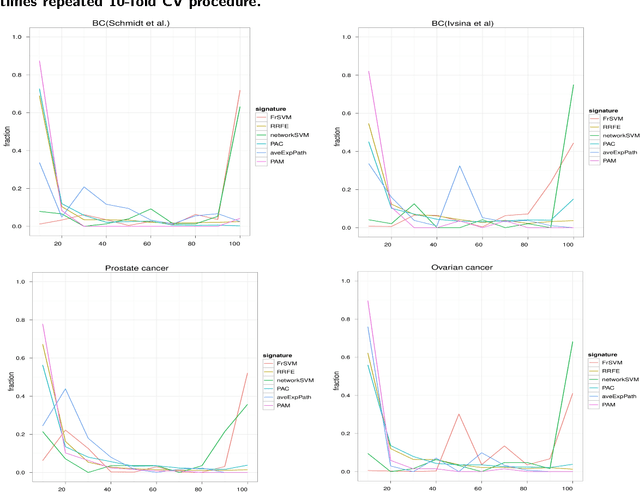
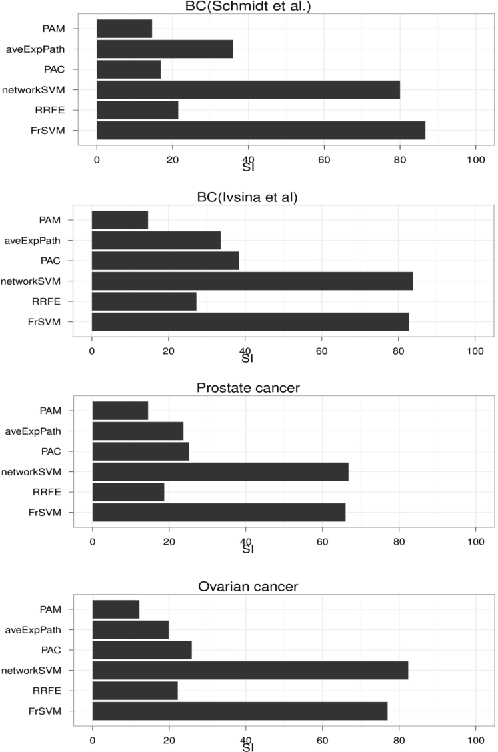
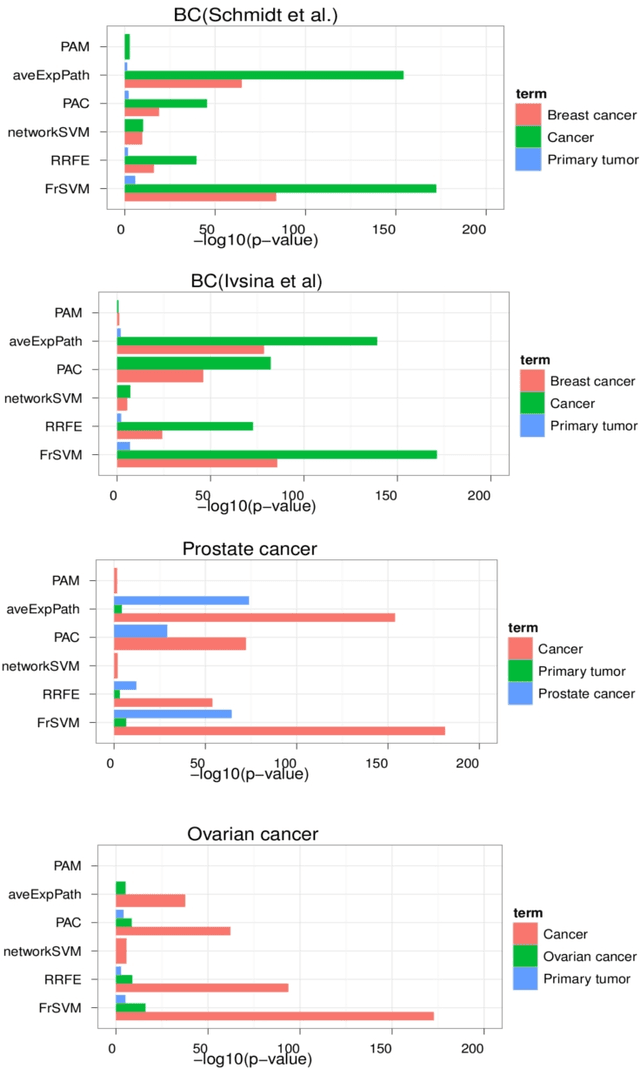
Abstract:Background: Predictive, stable and interpretable gene signatures are generally seen as an important step towards a better personalized medicine. During the last decade various methods have been proposed for that purpose. However, one important obstacle for making gene signatures a standard tool in clinics is the typical low reproducibility of these signatures combined with the difficulty to achieve a clear biological interpretation. For that purpose in the last years there has been a growing interest in approaches that try to integrate information from molecular interaction networks. Results: We propose a novel algorithm, called FrSVM, which integrates protein-protein interaction network information into gene selection for prognostic biomarker discovery. Our method is a simple filter based approach, which focuses on central genes with large differences in their expression. Compared to several other competing methods our algorithm reveals a significantly better prediction performance and higher signature stability. More- over, obtained gene lists are highly enriched with known disease genes and drug targets. We extendd our approach further by integrating information on candidate disease genes and targets of disease associated Transcript Factors (TFs).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge