Joseph Bockhorst
Graph Neural Networks to Predict Customer Satisfaction Following Interactions with a Corporate Call Center
Jan 31, 2021


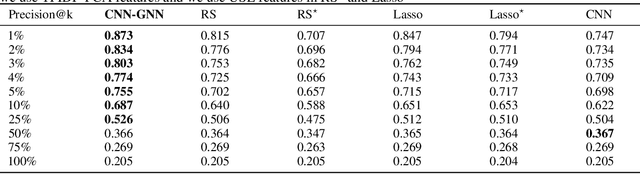
Abstract:Customer satisfaction is an important factor in creating and maintaining long-term relationships with customers. Near real-time identification of potentially dissatisfied customers following phone calls can provide organizations the opportunity to take meaningful interventions and to foster ongoing customer satisfaction and loyalty. This work describes a fully operational system we have developed at a large US company for predicting customer satisfaction following incoming phone calls. The system takes as an input speech-to-text transcriptions of calls and predicts call satisfaction reported by customers on post-call surveys (scale from 1 to 10). Because of its ordinal, subjective, and often highly-skewed nature, predicting survey scores is not a trivial task and presents several modeling challenges. We introduce a graph neural network (GNN) approach that takes into account the comparative nature of the problem by considering the relative scores among batches, instead of only pairs of calls when training. This approach produces more accurate predictions than previous approaches including standard regression and classification models that directly fit the survey scores with call data. Our proposed approach can be easily generalized to other customer satisfaction prediction problems.
Discovering Patterns in Biological Sequences by Optimal Segmentation
Jun 20, 2012



Abstract:Computational methods for discovering patterns of local correlations in sequences are important in computational biology. Here we show how to determine the optimal partitioning of aligned sequences into non-overlapping segments such that positions in the same segment are strongly correlated while positions in different segments are not. Our approach involves discovering the hidden variables of a Bayesian network that interact with observed sequences so as to form a set of independent mixture models. We introduce a dynamic program to efficiently discover the optimal segmentation, or equivalently the optimal set of hidden variables. We evaluate our approach on two computational biology tasks. One task is related to the design of vaccines against polymorphic pathogens and the other task involves analysis of single nucleotide polymorphisms (SNPs) in human DNA. We show how common tasks in these problems naturally correspond to inference procedures in the learned models. Error rates of our learned models for the prediction of missing SNPs are up to 1/3 less than the error rates of a state-of-the-art SNP prediction method. Source code is available at www.uwm.edu/~joebock/segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge