Joohyeok Kim
Rethinking Glaucoma Calibration: Voting-Based Binocular and Metadata Integration
Mar 24, 2025Abstract:Glaucoma is an incurable ophthalmic disease that damages the optic nerve, leads to vision loss, and ranks among the leading causes of blindness worldwide. Diagnosing glaucoma typically involves fundus photography, optical coherence tomography (OCT), and visual field testing. However, the high cost of OCT often leads to reliance on fundus photography and visual field testing, both of which exhibit inherent inter-observer variability. This stems from glaucoma being a multifaceted disease that influenced by various factors. As a result, glaucoma diagnosis is highly subjective, emphasizing the necessity of calibration, which aligns predicted probabilities with actual disease likelihood. Proper calibration is essential to prevent overdiagnosis or misdiagnosis, which are critical concerns for high-risk diseases. Although AI has significantly improved diagnostic accuracy, overconfidence in models have worsen calibration performance. Recent study has begun focusing on calibration for glaucoma. Nevertheless, previous study has not fully considered glaucoma's systemic nature and the high subjectivity in its diagnostic process. To overcome these limitations, we propose V-ViT (Voting-based ViT), a novel framework that enhances calibration by incorporating disease-specific characteristics. V-ViT integrates binocular data and metadata, reflecting the multi-faceted nature of glaucoma diagnosis. Additionally, we introduce a MC dropout-based Voting System to address high subjectivity. Our approach achieves state-of-the-art performance across all metrics, including accuracy, demonstrating that our proposed methods are effective in addressing calibration issues. We validate our method using a custom dataset including binocular data.
EP-SAM: Weakly Supervised Histopathology Segmentation via Enhanced Prompt with Segment Anything
Oct 22, 2024
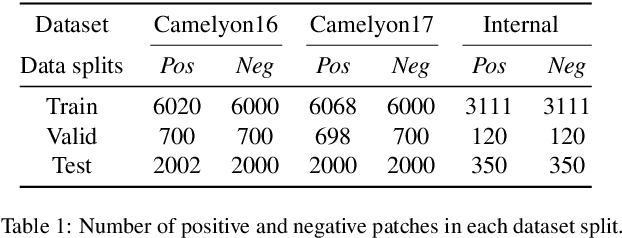
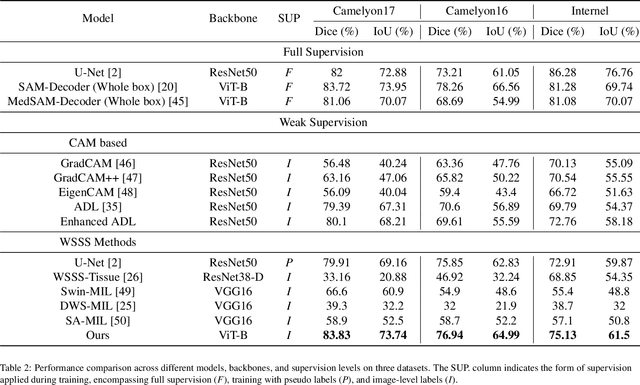

Abstract:This work proposes a novel approach beyond supervised learning for effective pathological image analysis, addressing the challenge of limited robust labeled data. Pathological diagnosis of diseases like cancer has conventionally relied on the evaluation of morphological features by physicians and pathologists. However, recent advancements in compute-aided diagnosis (CAD) systems are gaining significant attention as diagnostic support tools. Although the advancement of deep learning has improved CAD significantly, segmentation models typically require large pixel-level annotated dataset, and such labeling is expensive. Existing studies not based on supervised approaches still struggle with limited generalization, and no practical approach has emerged yet. To address this issue, we present a weakly supervised semantic segmentation (WSSS) model by combining class activation map and Segment Anything Model (SAM)-based pseudo-labeling. For effective pretraining, we adopt the SAM-a foundation model that is pretrained on large datasets and operates in zero-shot configurations using only coarse prompts. The proposed approach transfer enhanced Attention Dropout Layer's knowledge to SAM, thereby generating pseudo-labels. To demonstrate the superiority of the proposed method, experimental studies are conducted on histopathological breast cancer datasets. The proposed method outperformed other WSSS methods across three datasets, demonstrating its efficiency by achieving this with only 12GB of GPU memory during training. Our code is available at : https://github.com/QI-NemoSong/EP-SAM
Enhanced Prompt-leveraged Weakly Supervised Cancer Segmentation based on Segment Anything
Oct 17, 2024



Abstract:This work proposes a novel approach beyond supervised learning for effective pathological image analysis, addressing the challenge of limited robust labeled data. Pathological diagnosis of diseases like cancer has conventionally relied on the evaluation of morphological features by physicians and pathologists. However, recent advancements in compute-aided diagnosis (CAD) systems are gaining significant attention as diagnostic support tools. Although the advancement of deep learning has improved CAD significantly, segmentation models typically require large pixel-level annotated dataset, and such labeling is expensive. Existing studies not based on supervised approaches still struggle with limited generalization, and no practical approach has emerged yet. To address this issue, we present a weakly supervised semantic segmentation (WSSS) model by combining class activation map and Segment Anything Model (SAM)-based pseudo-labeling. For effective pretraining, we adopt the SAM-a foundation model that is pretrained on large datasets and operates in zero-shot configurations using only coarse prompts. The proposed approach transfer enhanced Attention Dropout Layer's knowledge to SAM, thereby generating pseudo-labels. To demonstrate the superiority of the proposed method, experimental studies are conducted on histopathological breast cancer datasets. The proposed method outperformed other WSSS methods across three datasets, demonstrating its efficiency by achieving this with only 12GB of GPU memory during training. Our code is available at : https://github.com/QI-NemoSong/EPLC-SAM
CAD: Memory Efficient Convolutional Adapter for Segment Anything
Sep 24, 2024



Abstract:The Foundation model for image segmentation, Segment Anything (SAM), has been actively researched in various fields since its proposal. Various researches have been proposed to adapt SAM to specific domains, with one notable approach involving the addition and training of lightweight adapter modules. While adapter-based fine-tuning approaches have reported parameter efficiency and significant performance improvements, they face a often overlooked issue: the excessive consumption of GPU memory relative to the number of trainable parameters. Addressing this issue, this paper proposes a memory-efficient parallel convolutional adapter architecture. This architecture connects in parallel with SAM's image encoder, eliminating the need to store activations and gradients of the image encoder during model training. Our proposed architecture demonstrated competitive experimental results while using less than half the GPU memory compared to SAM Adapter, indicating its value as an alternative to simple decoder fine-tuning when hardware limitations preclude adapter-based learning. Our code implementation is available at our github.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge