Jonathan Campbell
Exploring Levels of Control for a Navigation Assistant for Blind Travelers
Jan 05, 2023
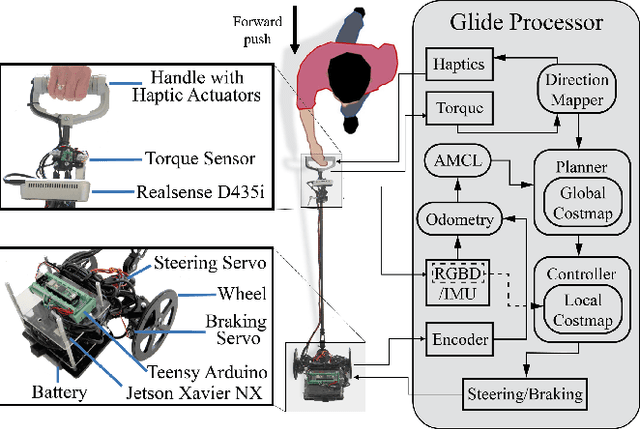
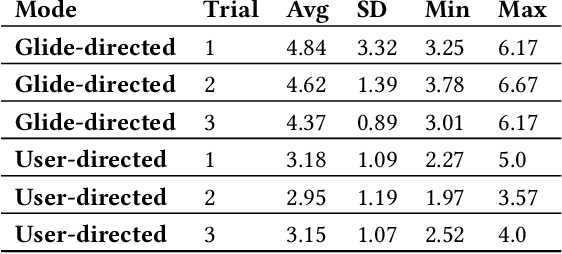

Abstract:Only a small percentage of blind and low-vision people use traditional mobility aids such as a cane or a guide dog. Various assistive technologies have been proposed to address the limitations of traditional mobility aids. These devices often give either the user or the device majority of the control. In this work, we explore how varying levels of control affect the users' sense of agency, trust in the device, confidence, and successful navigation. We present Glide, a novel mobility aid with two modes for control: Glide-directed and User-directed. We employ Glide in a study (N=9) in which blind or low-vision participants used both modes to navigate through an indoor environment. Overall, participants found that Glide was easy to use and learn. Most participants trusted Glide despite its current limitations, and their confidence and performance increased as they continued to use Glide. Users' control mode preference varied in different situations; no single mode "won" in all situations.
A New Graph Node Classification Benchmark: Learning Structure from Histology Cell Graphs
Nov 11, 2022

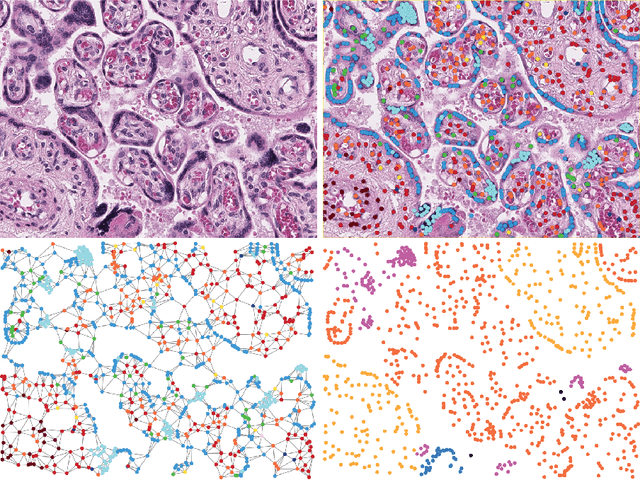

Abstract:We introduce a new benchmark dataset, Placenta, for node classification in an underexplored domain: predicting microanatomical tissue structures from cell graphs in placenta histology whole slide images. This problem is uniquely challenging for graph learning for a few reasons. Cell graphs are large (>1 million nodes per image), node features are varied (64-dimensions of 11 types of cells), class labels are imbalanced (9 classes ranging from 0.21% of the data to 40.0%), and cellular communities cluster into heterogeneously distributed tissues of widely varying sizes (from 11 nodes to 44,671 nodes for a single structure). Here, we release a dataset consisting of two cell graphs from two placenta histology images totalling 2,395,747 nodes, 799,745 of which have ground truth labels. We present inductive benchmark results for 7 scalable models and show how the unique qualities of cell graphs can help drive the development of novel graph neural network architectures.
Swarming around Shellfish Larvae
Dec 17, 2004



Abstract:The collection of wild larvae seed as a source of raw material is a major sub industry of shellfish aquaculture. To predict when, where and in what quantities wild seed will be available, it is necessary to track the appearance and growth of planktonic larvae. One of the most difficult groups to identify, particularly at the species level are the Bivalvia. This difficulty arises from the fact that fundamentally all bivalve larvae have a similar shape and colour. Identification based on gross morphological appearance is limited by the time-consuming nature of the microscopic examination and by the limited availability of expertise in this field. Molecular and immunological methods are also being studied. We describe the application of computational pattern recognition methods to the automated identification and size analysis of scallop larvae. For identification, the shape features used are binary invariant moments; that is, the features are invariant to shift (position within the image), scale (induced either by growth or differential image magnification) and rotation. Images of a sample of scallop and non-scallop larvae covering a range of maturities have been analysed. In order to overcome the automatic identification, as well as to allow the system to receive new unknown samples at any moment, a self-organized and unsupervised ant-like clustering algorithm based on Swarm Intelligence is proposed, followed by simple k-NNR nearest neighbour classification on the final map. Results achieve a full recognition rate of 100% under several situations (k =1 or 3).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge