John Scholefield
An ensemble of machine learning and anti-learning methods for predicting tumour patient survival rates
Jul 21, 2016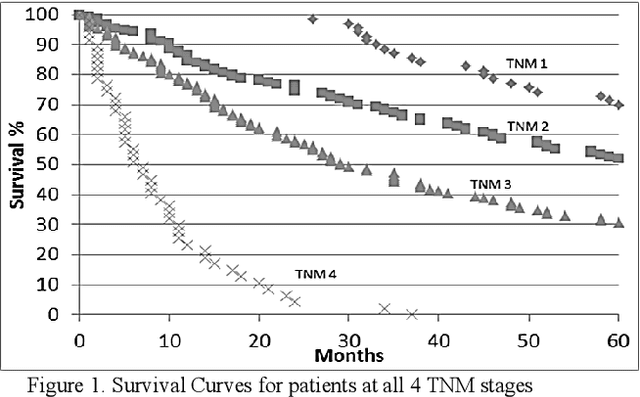

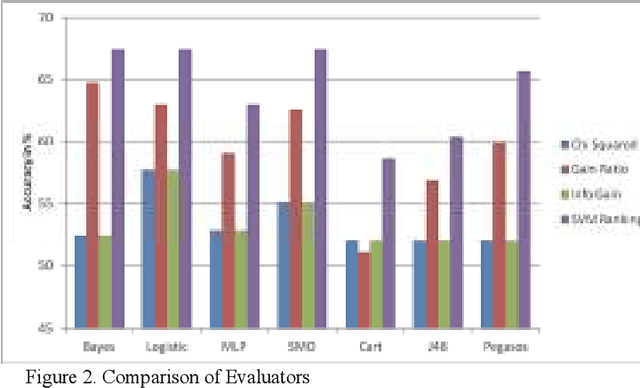
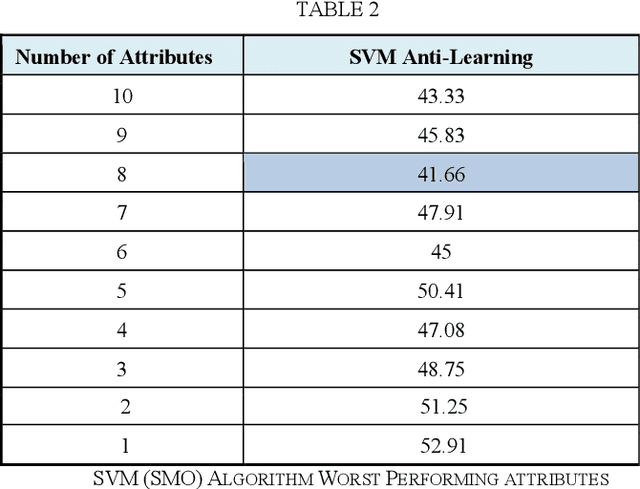
Abstract:This paper primarily addresses a dataset relating to cellular, chemical and physical conditions of patients gathered at the time they are operated upon to remove colorectal tumours. This data provides a unique insight into the biochemical and immunological status of patients at the point of tumour removal along with information about tumour classification and post-operative survival. The relationship between severity of tumour, based on TNM staging, and survival is still unclear for patients with TNM stage 2 and 3 tumours. We ask whether it is possible to predict survival rate more accurately using a selection of machine learning techniques applied to subsets of data to gain a deeper understanding of the relationships between a patient's biochemical markers and survival. We use a range of feature selection and single classification techniques to predict the 5 year survival rate of TNM stage 2 and 3 patients which initially produces less than ideal results. The performance of each model individually is then compared with subsets of the data where agreement is reached for multiple models. This novel method of selective ensembling demonstrates that significant improvements in model accuracy on an unseen test set can be achieved for patients where agreement between models is achieved. Finally we point at a possible method to identify whether a patients prognosis can be accurately predicted or not.
Ensemble Learning of Colorectal Cancer Survival Rates
Sep 02, 2014
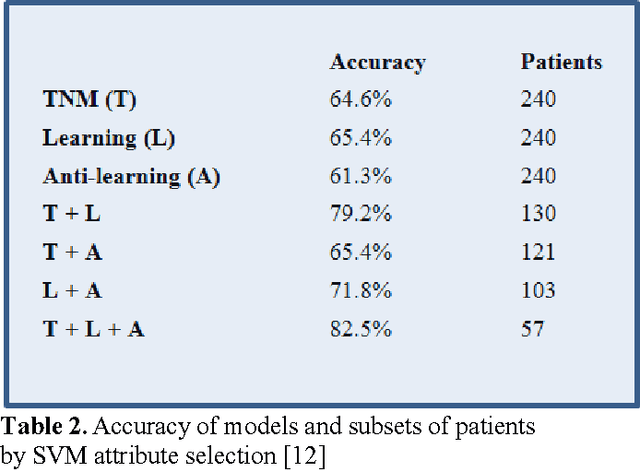

Abstract:In this paper, we describe a dataset relating to cellular and physical conditions of patients who are operated upon to remove colorectal tumours. This data provides a unique insight into immunological status at the point of tumour removal, tumour classification and post-operative survival. We build on existing research on clustering and machine learning facets of this data to demonstrate a role for an ensemble approach to highlighting patients with clearer prognosis parameters. Results for survival prediction using 3 different approaches are shown for a subset of the data which is most difficult to model. The performance of each model individually is compared with subsets of the data where some agreement is reached for multiple models. Significant improvements in model accuracy on an unseen test set can be achieved for patients where agreement between models is achieved.
Biomarker Clustering of Colorectal Cancer Data to Complement Clinical Classification
Jul 05, 2013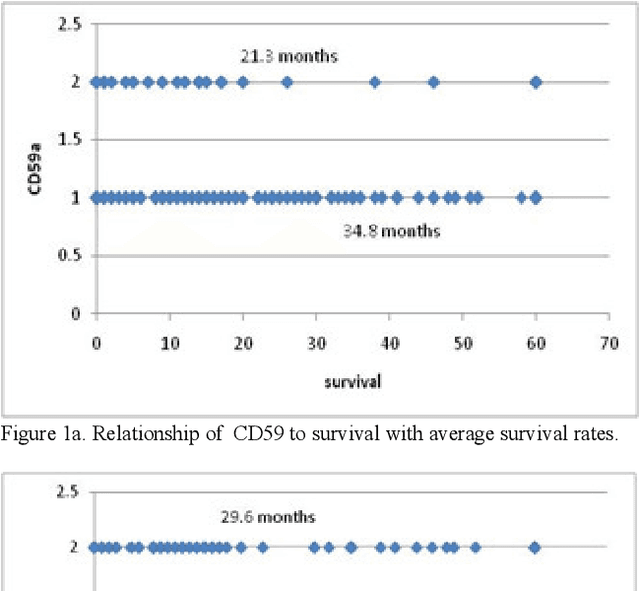
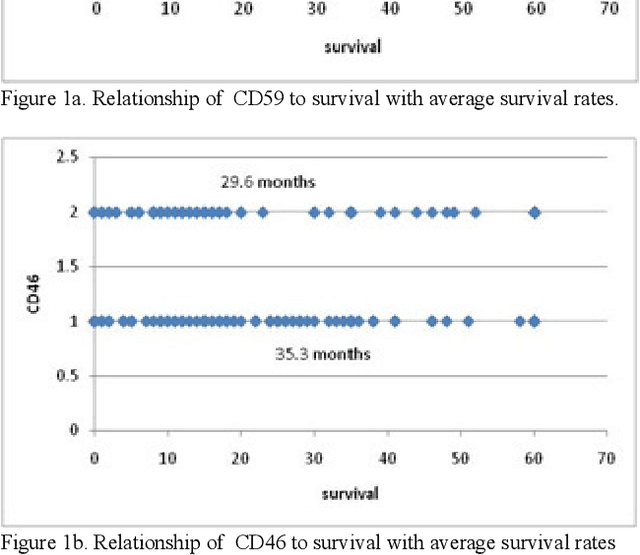
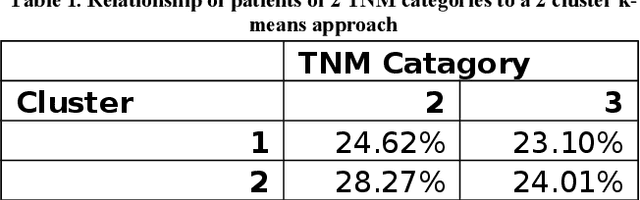

Abstract:In this paper, we describe a dataset relating to cellular and physical conditions of patients who are operated upon to remove colorectal tumours. This data provides a unique insight into immunological status at the point of tumour removal, tumour classification and post-operative survival. Attempts are made to cluster this dataset and important subsets of it in an effort to characterize the data and validate existing standards for tumour classification. It is apparent from optimal clustering that existing tumour classification is largely unrelated to immunological factors within a patient and that there may be scope for re-evaluating treatment options and survival estimates based on a combination of tumour physiology and patient histochemistry.
Supervised Learning and Anti-learning of Colorectal Cancer Classes and Survival Rates from Cellular Biology Parameters
Jul 05, 2013
Abstract:In this paper, we describe a dataset relating to cellular and physical conditions of patients who are operated upon to remove colorectal tumours. This data provides a unique insight into immunological status at the point of tumour removal, tumour classification and post-operative survival. Attempts are made to learn relationships between attributes (physical and immunological) and the resulting tumour stage and survival. Results for conventional machine learning approaches can be considered poor, especially for predicting tumour stages for the most important types of cancer. This poor performance is further investigated and compared with a synthetic, dataset based on the logical exclusive-OR function and it is shown that there is a significant level of 'anti-learning' present in all supervised methods used and this can be explained by the highly dimensional, complex and sparsely representative dataset. For predicting the stage of cancer from the immunological attributes, anti-learning approaches outperform a range of popular algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge