Jingwei Qu
DS-HGCN: A Dual-Stream Hypergraph Convolutional Network for Predicting Student Engagement via Social Contagion
Dec 23, 2025Abstract:Student engagement is a critical factor influencing academic success and learning outcomes. Accurately predicting student engagement is essential for optimizing teaching strategies and providing personalized interventions. However, most approaches focus on single-dimensional feature analysis and assessing engagement based on individual student factors. In this work, we propose a dual-stream multi-feature fusion model based on hypergraph convolutional networks (DS-HGCN), incorporating social contagion of student engagement. DS-HGCN enables accurate prediction of student engagement states by modeling multi-dimensional features and their propagation mechanisms between students. The framework constructs a hypergraph structure to encode engagement contagion among students and captures the emotional and behavioral differences and commonalities by multi-frequency signals. Furthermore, we introduce a hypergraph attention mechanism to dynamically weigh the influence of each student, accounting for individual differences in the propagation process. Extensive experiments on public benchmark datasets demonstrate that our proposed method achieves superior performance and significantly outperforms existing state-of-the-art approaches.
Multi-Prototype Embedding Refinement for Semi-Supervised Medical Image Segmentation
Mar 18, 2025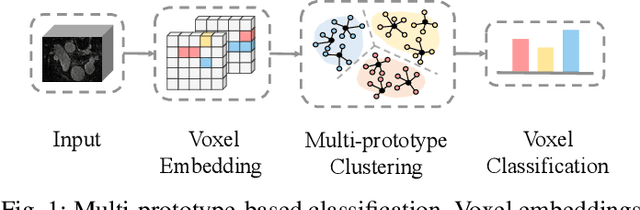
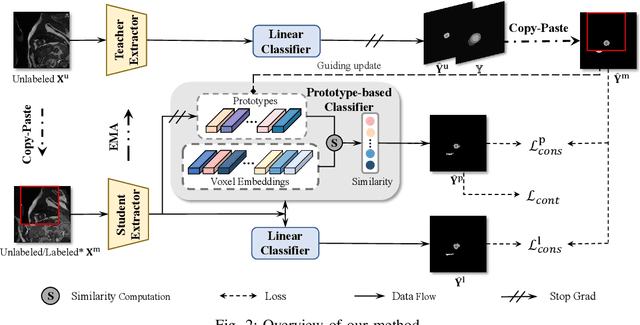
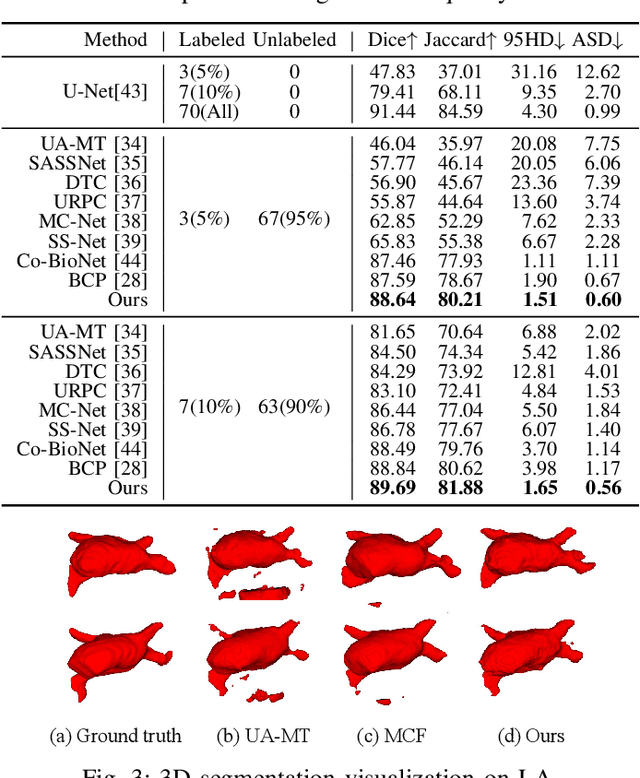
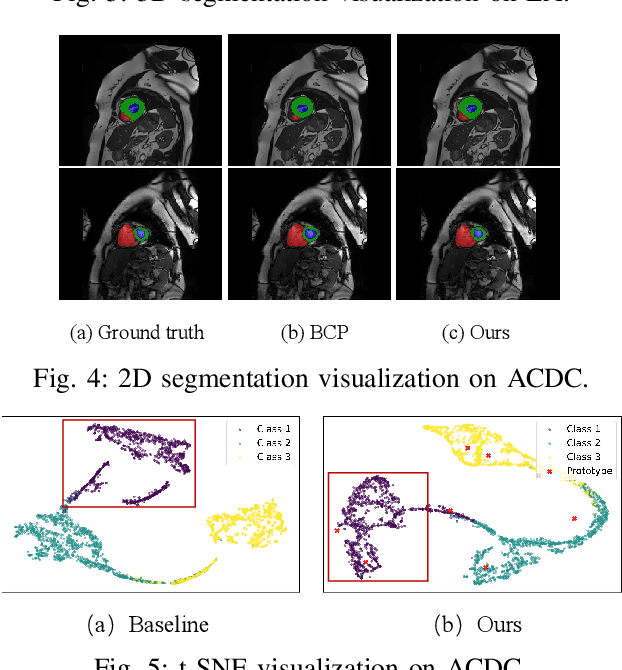
Abstract:Medical image segmentation aims to identify anatomical structures at the voxel-level. Segmentation accuracy relies on distinguishing voxel differences. Compared to advancements achieved in studies of the inter-class variance, the intra-class variance receives less attention. Moreover, traditional linear classifiers, limited by a single learnable weight per class, struggle to capture this finer distinction. To address the above challenges, we propose a Multi-Prototype-based Embedding Refinement method for semi-supervised medical image segmentation. Specifically, we design a multi-prototype-based classification strategy, rethinking the segmentation from the perspective of structural relationships between voxel embeddings. The intra-class variations are explored by clustering voxels along the distribution of multiple prototypes in each class. Next, we introduce a consistency constraint to alleviate the limitation of linear classifiers. This constraint integrates different classification granularities from a linear classifier and the proposed prototype-based classifier. In the thorough evaluation on two popular benchmarks, our method achieves superior performance compared with state-of-the-art methods. Code is available at https://github.com/Briley-byl123/MPER.
Graph Attribute Aggregation Network with Progressive Margin Folding
May 14, 2019



Abstract:Graph convolutional neural networks (GCNNs) have been attracting increasing research attention due to its great potential in inference over graph structures. However, insufficient effort has been devoted to the aggregation methods between different convolution graph layers. In this paper, we introduce a graph attribute aggregation network (GAAN) architecture. Different from the conventional pooling operations, a graph-transformation-based aggregation strategy, progressive margin folding, PMF, is proposed for integrating graph features. By distinguishing internal and margin elements, we provide an approach for implementing the folding iteratively. And a mechanism is also devised for preserving the local structures during progressively folding. In addition, a hypergraph-based representation is introduced for transferring the aggregated information between different layers. Our experiments applied to the public molecule datasets demonstrate that the proposed GAAN outperforms the existing GCNN models with significant effectiveness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge