Jin-Dong Kim
Knowledge-Driven Mechanistic Enrichment of the Preeclampsia Ignorome
Jul 28, 2022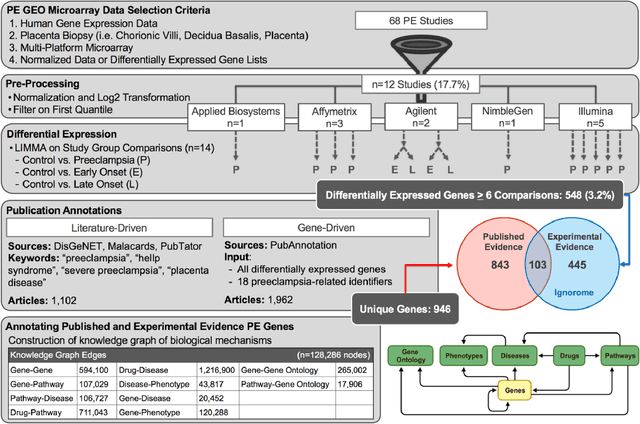
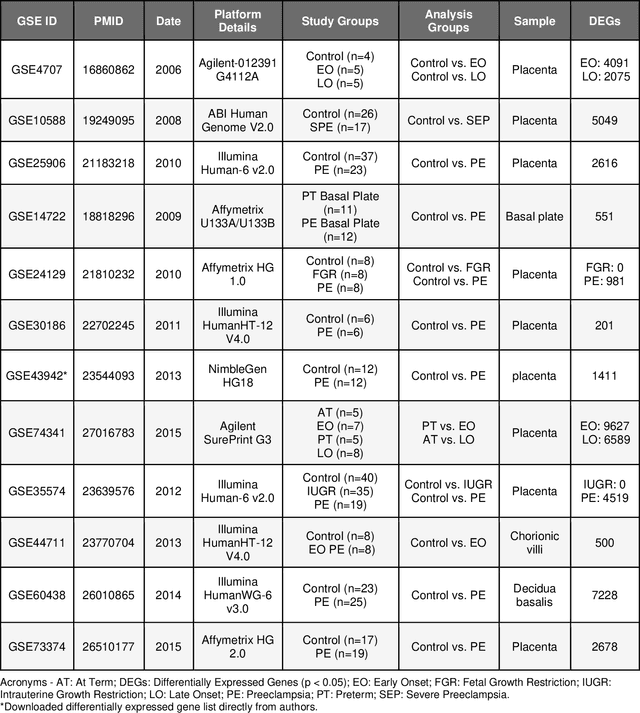
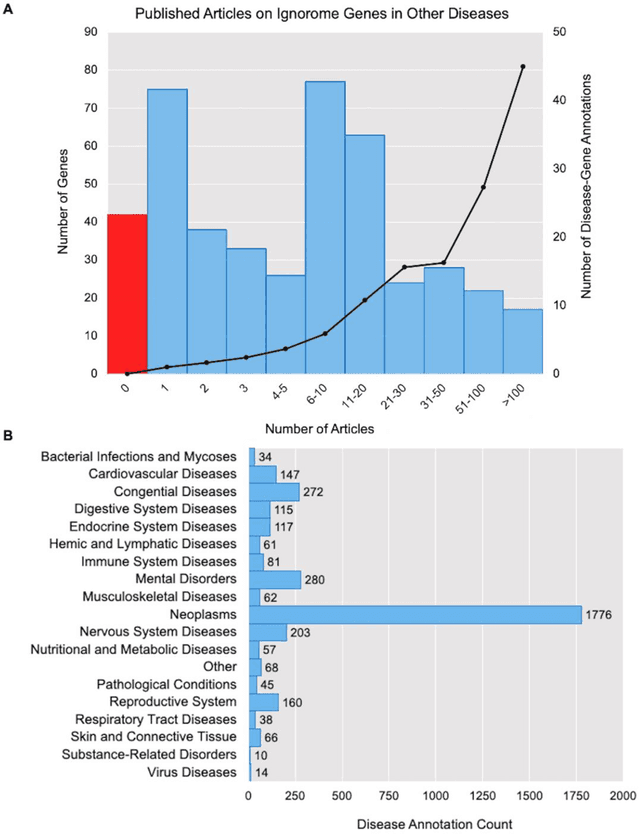
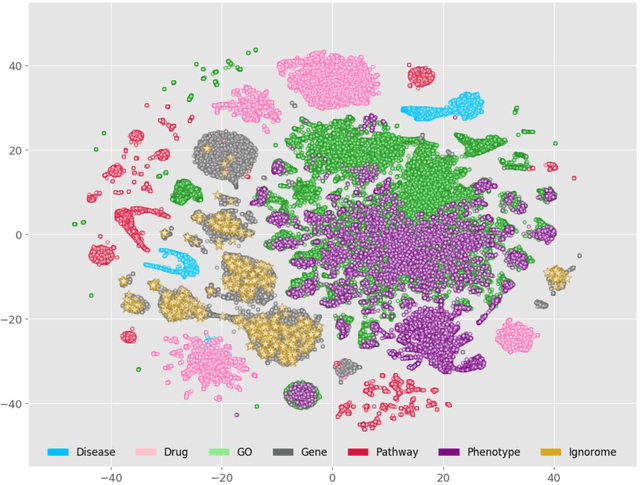
Abstract:Preeclampsia is a leading cause of maternal and fetal morbidity and mortality. Currently, the only definitive treatment of preeclampsia is delivery of the placenta, which is central to the pathogenesis of the disease. Transcriptional profiling of human placenta from pregnancies complicated by preeclampsia has been extensively performed to identify differentially expressed genes (DEGs). DEGs are identified using unbiased assays, however, the decisions to investigate DEGs experimentally are biased by many factors, causing many DEGs to remain uninvestigated. A set of DEGs which are associated with a disease experimentally, but which have no known association with the disease in the literature is known as the ignorome. Preeclampsia has an extensive body of scientific literature, a large pool of DEG data, and only one definitive treatment. Tools facilitating knowledge-based analyses, which are capable of combining disparate data from many sources in order to suggest underlying mechanisms of action, may be a valuable resource to support discovery and improve our understanding of this disease. In this work we demonstrate how a biomedical knowledge graph (KG) can be used to identify novel preeclampsia molecular mechanisms. Existing open source biomedical resources and publicly available high-throughput transcriptional profiling data were used to identify and annotate the function of currently uninvestigated preeclampsia-associated DEGs. Experimentally investigated genes associated with preeclampsia were identified from PubMed abstracts using text-mining methodologies. The relative complement of the text-mined- and meta-analysis-derived lists were identified as the uninvestigated preeclampsia-associated DEGs (n=445), i.e., the preeclampsia ignorome. Using the KG to investigate relevant DEGs revealed 53 novel clinically relevant and biologically actionable mechanistic associations.
A Quantitative Evaluation of Natural Language Question Interpretation for Question Answering Systems
Sep 20, 2018
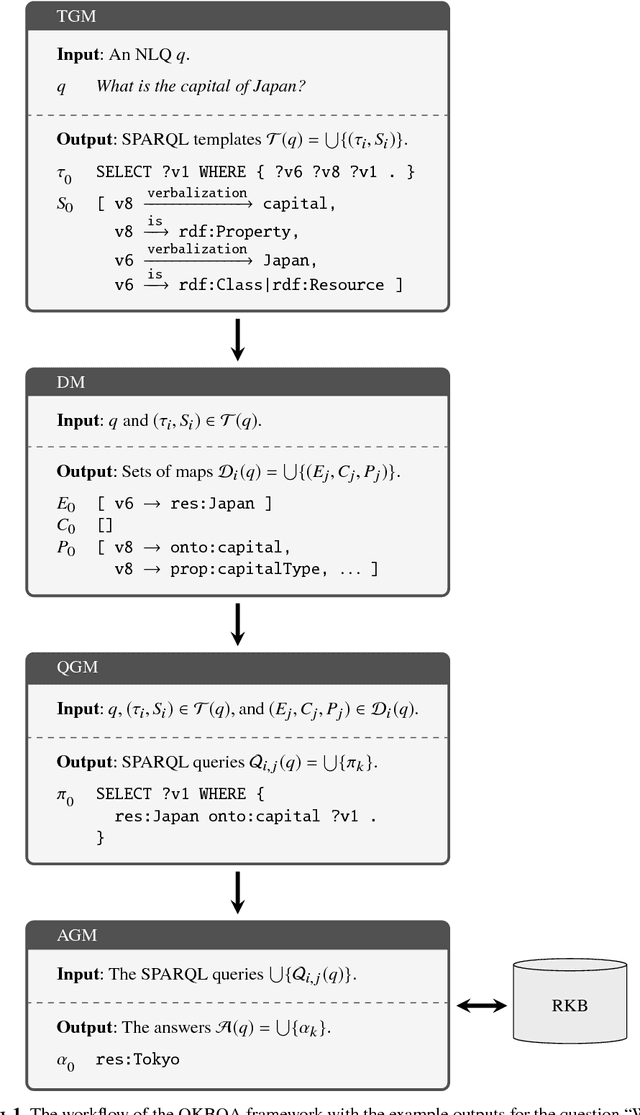
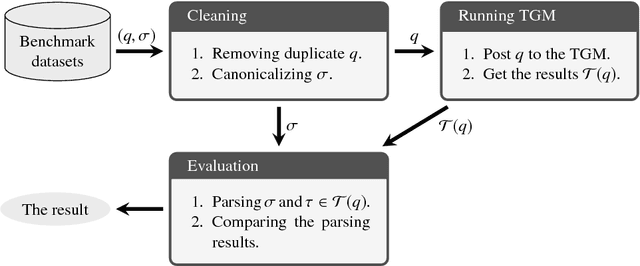

Abstract:Systematic benchmark evaluation plays an important role in the process of improving technologies for Question Answering (QA) systems. While currently there are a number of existing evaluation methods for natural language (NL) QA systems, most of them consider only the final answers, limiting their utility within a black box style evaluation. Herein, we propose a subdivided evaluation approach to enable finer-grained evaluation of QA systems, and present an evaluation tool which targets the NL question (NLQ) interpretation step, an initial step of a QA pipeline. The results of experiments using two public benchmark datasets suggest that we can get a deeper insight about the performance of a QA system using the proposed approach, which should provide a better guidance for improving the systems, than using black box style approaches.
HMM Specialization with Selective Lexicalization
Dec 23, 1999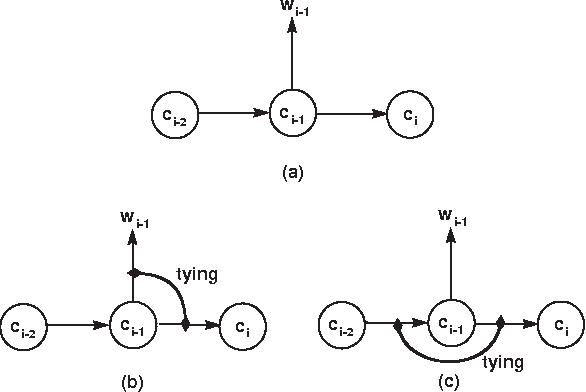
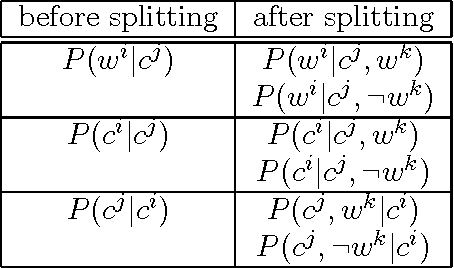
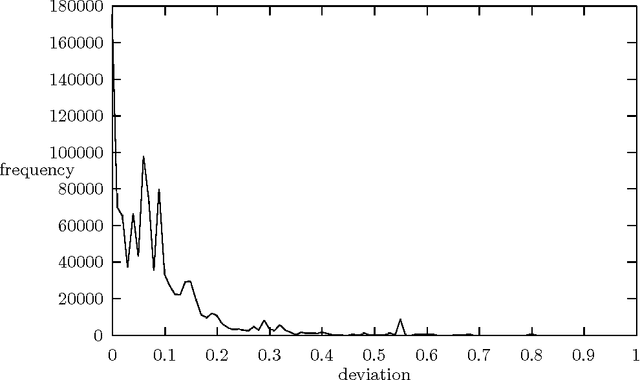
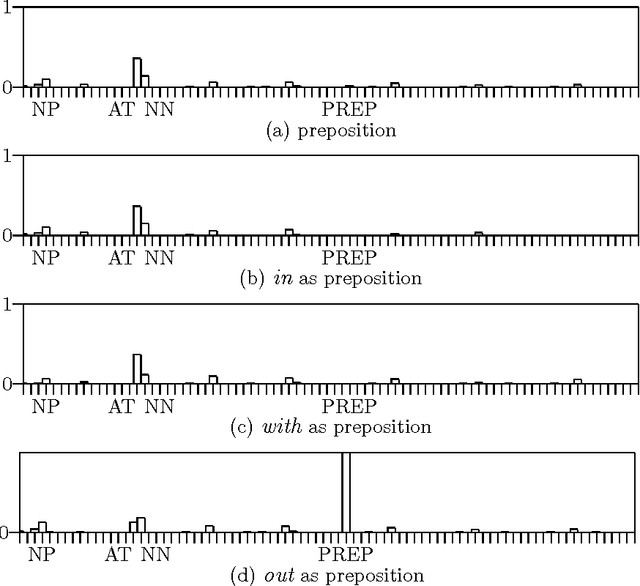
Abstract:We present a technique which complements Hidden Markov Models by incorporating some lexicalized states representing syntactically uncommon words. Our approach examines the distribution of transitions, selects the uncommon words, and makes lexicalized states for the words. We performed a part-of-speech tagging experiment on the Brown corpus to evaluate the resultant language model and discovered that this technique improved the tagging accuracy by 0.21% at the 95% level of confidence.
* 7 pages, 6 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge