Jiasheng Zhou
OmniCorpus: A Unified Multimodal Corpus of 10 Billion-Level Images Interleaved with Text
Jun 13, 2024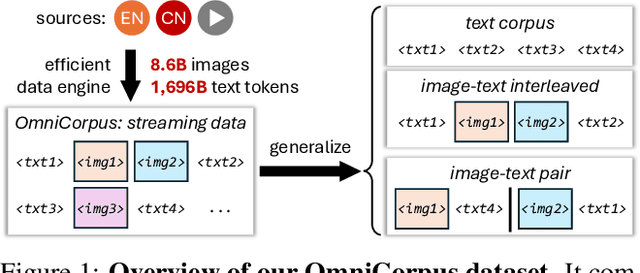
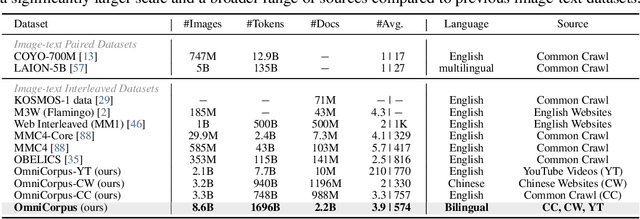

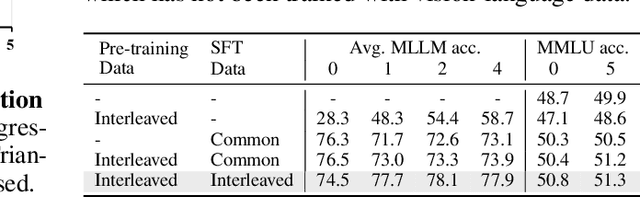
Abstract:Image-text interleaved data, consisting of multiple images and texts arranged in a natural document format, aligns with the presentation paradigm of internet data and closely resembles human reading habits. Recent studies have shown that such data aids multimodal in-context learning and maintains the capabilities of large language models during multimodal fine-tuning. However, the limited scale and diversity of current image-text interleaved data restrict the development of multimodal large language models. In this paper, we introduce OmniCorpus, a 10 billion-scale image-text interleaved dataset. Using an efficient data engine, we filter and extract large-scale high-quality documents, which contain 8.6 billion images and 1,696 billion text tokens. Compared to counterparts (e.g., MMC4, OBELICS), our dataset 1) has 15 times larger scales while maintaining good data quality; 2) features more diverse sources, including both English and non-English websites as well as video-centric websites; 3) is more flexible, easily degradable from an image-text interleaved format to pure text corpus and image-text pairs. Through comprehensive analysis and experiments, we validate the quality, usability, and effectiveness of the proposed dataset. We hope this could provide a solid data foundation for future multimodal model research. Code and data are released at https://github.com/OpenGVLab/OmniCorpus.
OmniCorpus: An Unified Multimodal Corpus of 10 Billion-Level Images Interleaved with Text
Jun 12, 2024



Abstract:Image-text interleaved data, consisting of multiple images and texts arranged in a natural document format, aligns with the presentation paradigm of internet data and closely resembles human reading habits. Recent studies have shown that such data aids multimodal in-context learning and maintains the capabilities of large language models during multimodal fine-tuning. However, the limited scale and diversity of current image-text interleaved data restrict the development of multimodal large language models. In this paper, we introduce OmniCorpus, a 10 billion-scale image-text interleaved dataset. Using an efficient data engine, we filter and extract large-scale high-quality documents, which contain 8.6 billion images and 1,696 billion text tokens. Compared to counterparts (e.g., MMC4, OBELICS), our dataset 1) has 15 times larger scales while maintaining good data quality; 2) features more diverse sources, including both English and non-English websites as well as video-centric websites; 3) is more flexible, easily degradable from an image-text interleaved format to pure text corpus and image-text pairs. Through comprehensive analysis and experiments, we validate the quality, usability, and effectiveness of the proposed dataset. We hope this could provide a solid data foundation for future multimodal model research. Code and data are released at https://github.com/OpenGVLab/OmniCorpus.
De-Noising of Photoacoustic Microscopy Images by Deep Learning
Jan 12, 2022



Abstract:As a hybrid imaging technology, photoacoustic microscopy (PAM) imaging suffers from noise due to the maximum permissible exposure of laser intensity, attenuation of ultrasound in the tissue, and the inherent noise of the transducer. De-noising is a post-processing method to reduce noise, and PAM image quality can be recovered. However, previous de-noising techniques usually heavily rely on mathematical priors as well as manually selected parameters, resulting in unsatisfactory and slow de-noising performance for different noisy images, which greatly hinders practical and clinical applications. In this work, we propose a deep learning-based method to remove complex noise from PAM images without mathematical priors and manual selection of settings for different input images. An attention enhanced generative adversarial network is used to extract image features and remove various noises. The proposed method is demonstrated on both synthetic and real datasets, including phantom (leaf veins) and in vivo (mouse ear blood vessels and zebrafish pigment) experiments. The results show that compared with previous PAM de-noising methods, our method exhibits good performance in recovering images qualitatively and quantitatively. In addition, the de-noising speed of 0.016 s is achieved for an image with $256\times256$ pixels. Our approach is effective and practical for the de-noising of PAM images.
Photoacoustic Microscopy with Sparse Data Enabled by Convolutional Neural Networks for Fast Imaging
Jun 08, 2020



Abstract:Photoacoustic microscopy (PAM) has been a promising biomedical imaging technology in recent years. However, the point-by-point scanning mechanism results in low-speed imaging, which limits the application of PAM. Reducing sampling density can naturally shorten image acquisition time, which is at the cost of image quality. In this work, we propose a method using convolutional neural networks (CNNs) to improve the quality of sparse PAM images, thereby speeding up image acquisition while keeping good image quality. The CNN model utilizes both squeeze-and-excitation blocks and residual blocks to achieve the enhancement, which is a mapping from a 1/4 or 1/16 low-sampling sparse PAM image to a latent fully-sampled image. The perceptual loss function is applied to keep the fidelity of images. The model is mainly trained and validated on PAM images of leaf veins. The experiments show the effectiveness of our proposed method, which significantly outperforms existing methods quantitatively and qualitatively. Our model is also tested using in vivo PAM images of blood vessels of mouse ears and eyes. The results show that the model can enhance the image quality of the sparse PAM image of blood vessels from several aspects, which may help fast PAM and facilitate its clinical applications.
Adaptive Weighting Depth-variant Deconvolution of Fluorescence Microscopy Images with Convolutional Neural Network
Jul 07, 2019



Abstract:Fluorescence microscopy plays an important role in biomedical research. The depth-variant point spread function (PSF) of a fluorescence microscope produces low-quality images especially in the out-of-focus regions of thick specimens. Traditional deconvolution to restore the out-of-focus images is usually insufficient since a depth-invariant PSF is assumed. This article aims at handling fluorescence microscopy images by learning-based depth-variant PSF and reducing artifacts. We propose adaptive weighting depth-variant deconvolution (AWDVD) with defocus level prediction convolutional neural network (DelpNet) to restore the out-of-focus images. Depth-variant PSFs of image patches can be obtained by DelpNet and applied in the afterward deconvolution. AWDVD is adopted for a whole image which is patch-wise deconvolved and appropriately cropped before deconvolution. DelpNet achieves the accuracy of 98.2%, which outperforms the best-ever one using the same microscopy dataset. Image patches of 11 defocus levels after deconvolution are validated with maximum improvement in the peak signal-to-noise ratio and structural similarity index of 6.6 dB and 11%, respectively. The adaptive weighting of the patch-wise deconvolved image can eliminate patch boundary artifacts and improve deconvolved image quality. The proposed method can accurately estimate depth-variant PSF and effectively recover out-of-focus microscopy images. To our acknowledge, this is the first study of handling out-of-focus microscopy images using learning-based depth-variant PSF. Facing one of the most common blurs in fluorescence microscopy, the novel method provides a practical technology to improve the image quality.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge