Jiabing Wang
Slice Imputation: Intermediate Slice Interpolation for Anisotropic 3D Medical Image Segmentation
Mar 21, 2022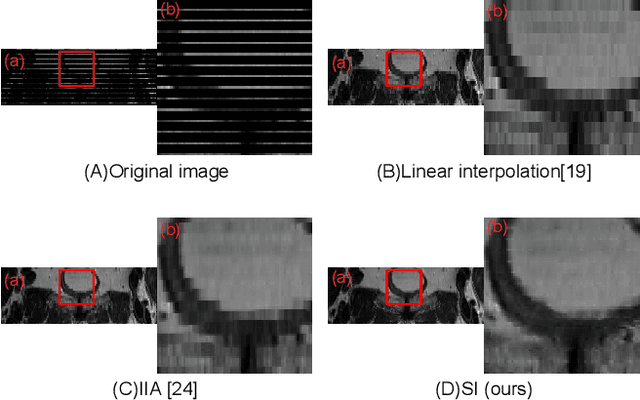
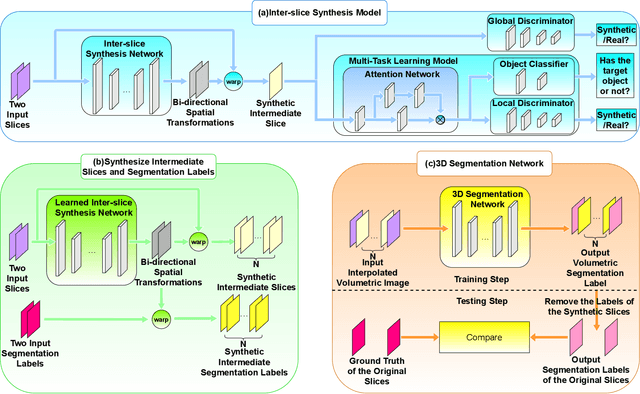
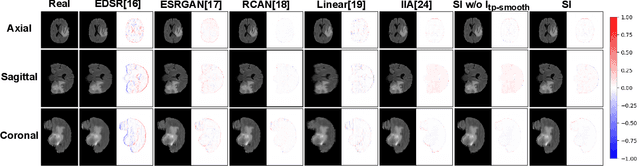
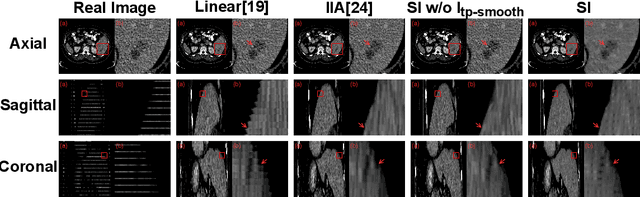
Abstract:We introduce a novel frame-interpolation-based method for slice imputation to improve segmentation accuracy for anisotropic 3D medical images, in which the number of slices and their corresponding segmentation labels can be increased between two consecutive slices in anisotropic 3D medical volumes. Unlike previous inter-slice imputation methods, which only focus on the smoothness in the axial direction, this study aims to improve the smoothness of the interpolated 3D medical volumes in all three directions: axial, sagittal, and coronal. The proposed multitask inter-slice imputation method, in particular, incorporates a smoothness loss function to evaluate the smoothness of the interpolated 3D medical volumes in the through-plane direction (sagittal and coronal). It not only improves the resolution of the interpolated 3D medical volumes in the through-plane direction but also transforms them into isotropic representations, which leads to better segmentation performances. Experiments on whole tumor segmentation in the brain, liver tumor segmentation, and prostate segmentation indicate that our method outperforms the competing slice imputation methods on both computed tomography and magnetic resonance images volumes in most cases.
Inter-slice image augmentation based on frame interpolation for boosting medical image segmentation accuracy
Jan 31, 2020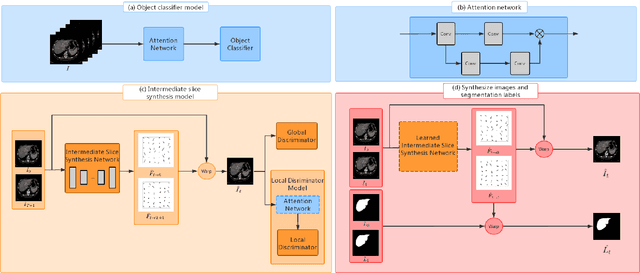
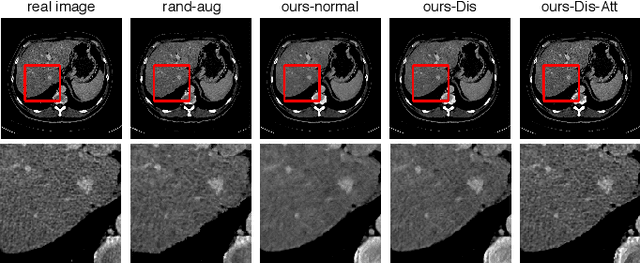
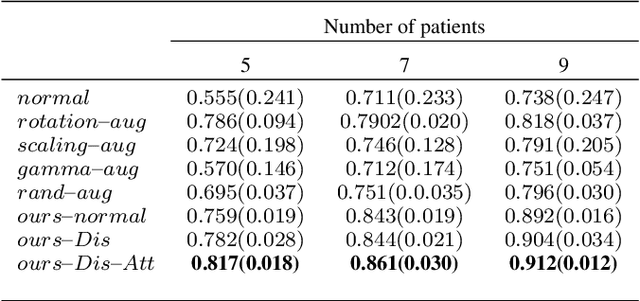
Abstract:We introduce the idea of inter-slice image augmentation whereby the numbers of the medical images and the corresponding segmentation labels are increased between two consecutive images in order to boost medical image segmentation accuracy. Unlike conventional data augmentation methods in medical imaging, which only increase the number of training samples directly by adding new virtual samples using simple parameterized transformations such as rotation, flipping, scaling, etc., we aim to augment data based on the relationship between two consecutive images, which increases not only the number but also the information of training samples. For this purpose, we propose a frame-interpolation-based data augmentation method to generate intermediate medical images and the corresponding segmentation labels between two consecutive images. We train and test a supervised U-Net liver segmentation network on SLIVER07 and CHAOS2019, respectively, with the augmented training samples, and obtain segmentation scores exhibiting significant improvement compared to the conventional augmentation methods.
Unified Attentional Generative Adversarial Network for Brain Tumor Segmentation From Multimodal Unpaired Images
Jul 08, 2019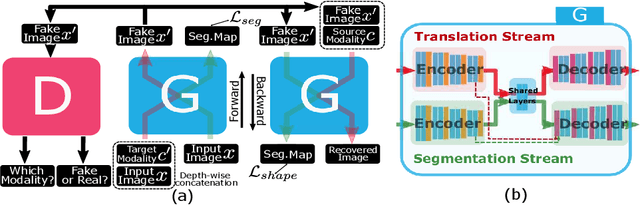
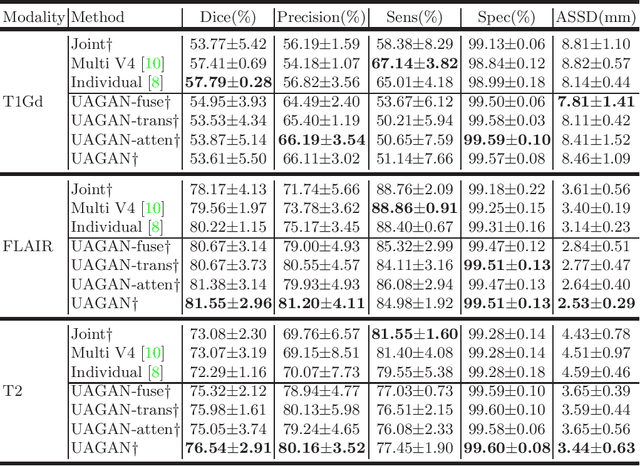
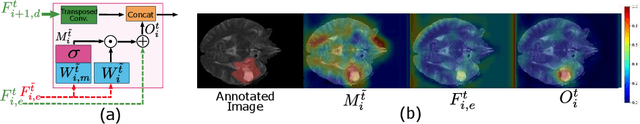
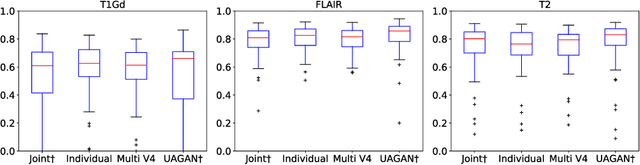
Abstract:In medical applications, the same anatomical structures may be observed in multiple modalities despite the different image characteristics. Currently, most deep models for multimodal segmentation rely on paired registered images. However, multimodal paired registered images are difficult to obtain in many cases. Therefore, developing a model that can segment the target objects from different modalities with unpaired images is significant for many clinical applications. In this work, we propose a novel two-stream translation and segmentation unified attentional generative adversarial network (UAGAN), which can perform any-to-any image modality translation and segment the target objects simultaneously in the case where two or more modalities are available. The translation stream is used to capture modality-invariant features of the target anatomical structures. In addition, to focus on segmentation-related features, we add attentional blocks to extract valuable features from the translation stream. Experiments on three-modality brain tumor segmentation indicate that UAGAN outperforms the existing methods in most cases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge