Jerry Chang
Feature-enhanced Adversarial Semi-supervised Semantic Segmentation Network for Pulmonary Embolism Annotation
Apr 08, 2022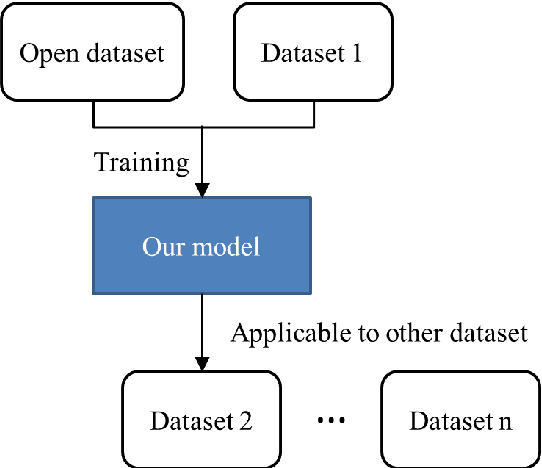

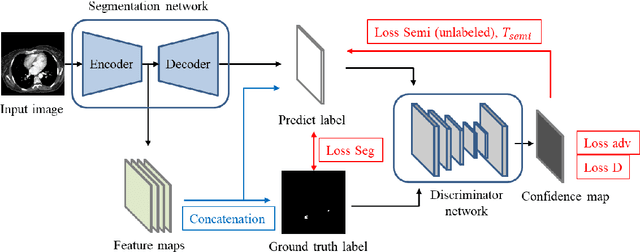
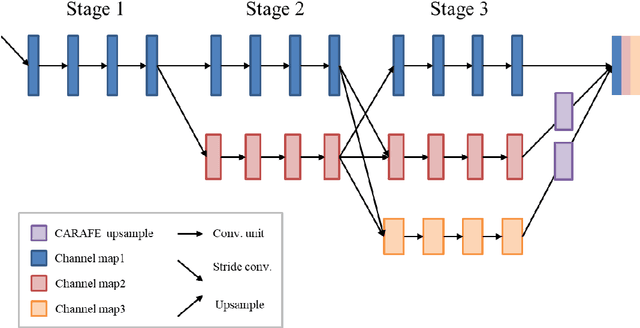
Abstract:This study established a feature-enhanced adversarial semi-supervised semantic segmentation model to automatically annotate pulmonary embolism lesion areas in computed tomography pulmonary angiogram (CTPA) images. In current studies, all of the PE CTPA image segmentation methods are trained by supervised learning. However, the supervised learning models need to be retrained and the images need to be relabeled when the CTPA images come from different hospitals. This study proposed a semi-supervised learning method to make the model applicable to different datasets by adding a small amount of unlabeled images. By training the model with both labeled and unlabeled images, the accuracy of unlabeled images can be improved and the labeling cost can be reduced. Our semi-supervised segmentation model includes a segmentation network and a discriminator network. We added feature information generated from the encoder of segmentation network to the discriminator so that it can learn the similarity between predicted mask and ground truth mask. This HRNet-based architecture can maintain a higher resolution for convolutional operations so the prediction of small PE lesion areas can be improved. We used the labeled open-source dataset and the unlabeled National Cheng Kung University Hospital (NCKUH) (IRB number: B-ER-108-380) dataset to train the semi-supervised learning model, and the resulting mean intersection over union (mIOU), dice score, and sensitivity achieved 0.3510, 0.4854, and 0.4253, respectively on the NCKUH dataset. Then, we fine-tuned and tested the model with a small amount of unlabeled PE CTPA images from China Medical University Hospital (CMUH) (IRB number: CMUH110-REC3-173) dataset. Comparing the results of our semi-supervised model with the supervised model, the mIOU, dice score, and sensitivity improved from 0.2344, 0.3325, and 0.3151 to 0.3721, 0.5113, and 0.4967, respectively.
The interpretation of endobronchial ultrasound image using 3D convolutional neural network for differentiating malignant and benign mediastinal lesions
Aug 02, 2021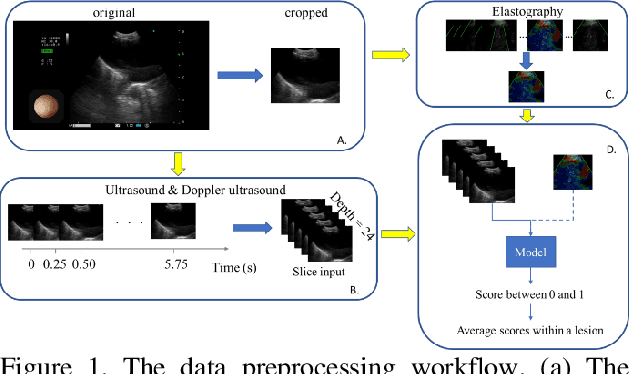
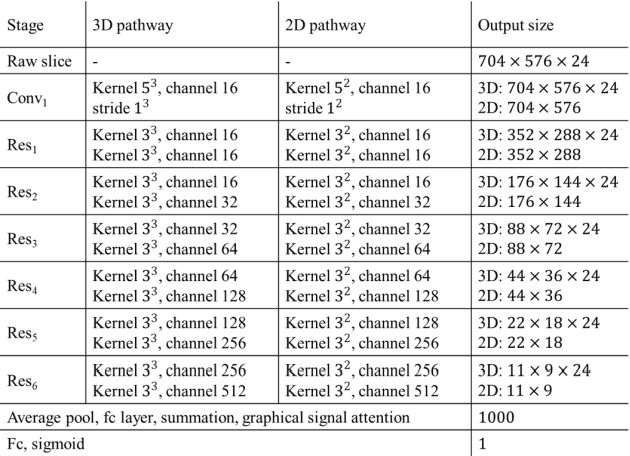
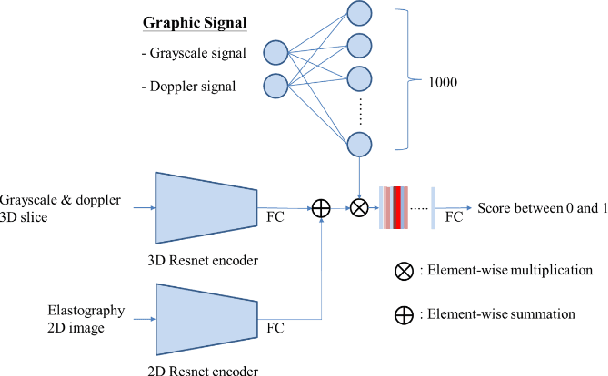
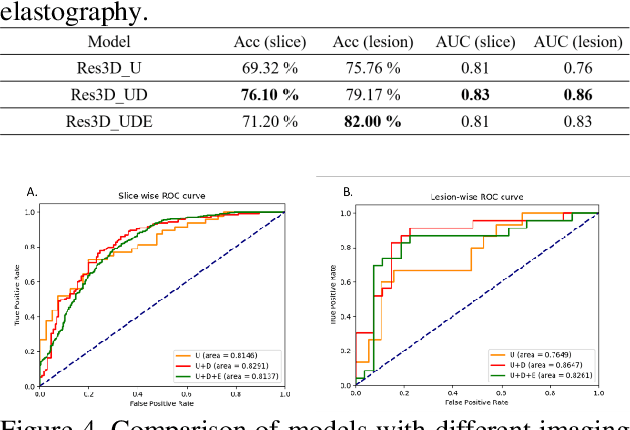
Abstract:The purpose of this study is to differentiate malignant and benign mediastinal lesions by using the three-dimensional convolutional neural network through the endobronchial ultrasound (EBUS) image. Compared with previous study, our proposed model is robust to noise and able to fuse various imaging features and spatiotemporal features of EBUS videos. Endobronchial ultrasound-guided transbronchial needle aspiration (EBUS-TBNA) is a diagnostic tool for intrathoracic lymph nodes. Physician can observe the characteristics of the lesion using grayscale mode, doppler mode, and elastography during the procedure. To process the EBUS data in the form of a video and appropriately integrate the features of multiple imaging modes, we used a time-series three-dimensional convolutional neural network (3D CNN) to learn the spatiotemporal features and design a variety of architectures to fuse each imaging mode. Our model (Res3D_UDE) took grayscale mode, Doppler mode, and elastography as training data and achieved an accuracy of 82.00% and area under the curve (AUC) of 0.83 on the validation set. Compared with previous study, we directly used videos recorded during procedure as training and validation data, without additional manual selection, which might be easier for clinical application. In addition, model designed with 3D CNN can also effectively learn spatiotemporal features and improve accuracy. In the future, our model may be used to guide physicians to quickly and correctly find the target lesions for slice sampling during the inspection process, reduce the number of slices of benign lesions, and shorten the inspection time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge