Ching-Kai Lin
Towards Enhanced Analysis of Lung Cancer Lesions in EBUS-TBNA -- A Semi-Supervised Video Object Detection Method
Apr 10, 2024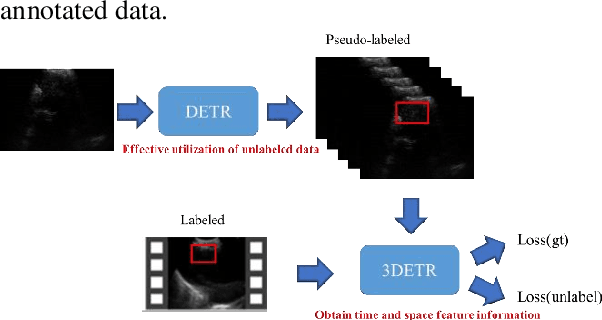
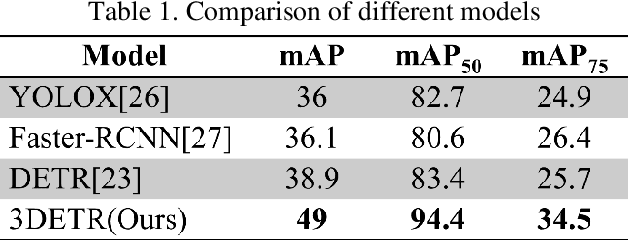
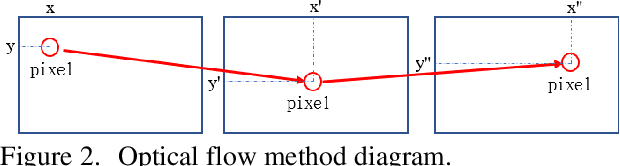
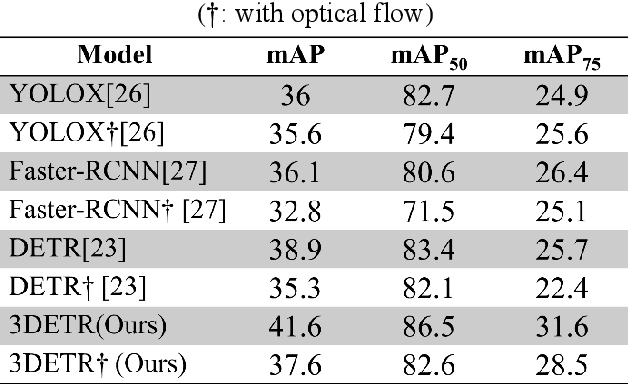
Abstract:This study aims to establish a computer-aided diagnostic system for lung lesions using bronchoscope endobronchial ultrasound (EBUS) to assist physicians in identifying lesion areas. During EBUS-transbronchial needle aspiration (EBUS-TBNA) procedures, physicians rely on grayscale ultrasound images to determine the location of lesions. However, these images often contain significant noise and can be influenced by surrounding tissues or blood vessels, making interpretation challenging. Previous research has lacked the application of object detection models to EBUS-TBNA, and there has been no well-defined solution for annotating the EBUS-TBNA dataset. In related studies on ultrasound images, although models have been successful in capturing target regions for their respective tasks, their training and predictions have been based on two-dimensional images, limiting their ability to leverage temporal features for improved predictions. This study introduces a three-dimensional image-based object detection model. It utilizes an attention mechanism to capture temporal correlations and we will implements a filtering mechanism to select relevant information from previous frames. Subsequently, a teacher-student model training approach is employed to optimize the model further, leveraging unlabeled data. To mitigate the impact of poor-quality pseudo-labels on the student model, we will add a special Gaussian Mixture Model (GMM) to ensure the quality of pseudo-labels.
Using Few-Shot Learning to Classify Primary Lung Cancer and Other Malignancy with Lung Metastasis in Cytological Imaging via Endobronchial Ultrasound Procedures
Apr 10, 2024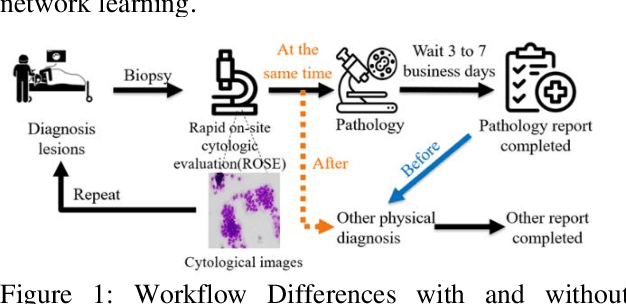
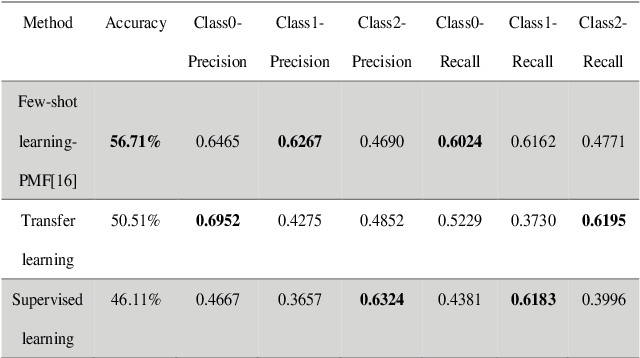
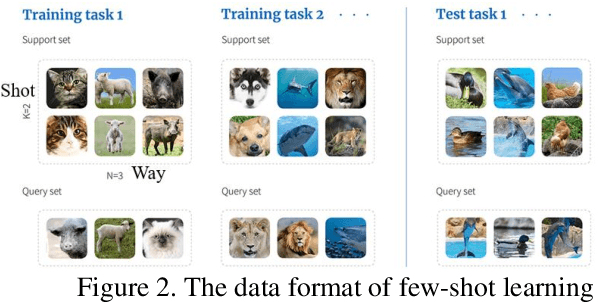
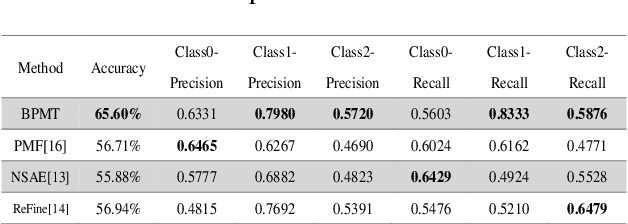
Abstract:This study aims to establish a computer-aided diagnosis system for endobronchial ultrasound (EBUS) surgery to assist physicians in the preliminary diagnosis of metastatic cancer. This involves arranging immediate examinations for other sites of metastatic cancer after EBUS surgery, eliminating the need to wait for reports, thereby shortening the waiting time by more than half and enabling patients to detect other cancers earlier, allowing for early planning and implementation of treatment plans. Unlike previous studies on cell image classification, which have abundant datasets for training, this study must also be able to make effective classifications despite the limited amount of case data for lung metastatic cancer. In the realm of small data set classification methods, Few-shot learning (FSL) has become mainstream in recent years. Through its ability to train on small datasets and its strong generalization capabilities, FSL shows potential in this task of lung metastatic cell image classification. This study will adopt the approach of Few-shot learning, referencing existing proposed models, and designing a model architecture for classifying lung metastases cell images. Batch Spectral Regularization (BSR) will be incorporated as a loss update parameter, and the Finetune method of PMF will be modified. In terms of test results, the addition of BSR and the modified Finetune method further increases the accuracy by 8.89% to 65.60%, outperforming other FSL methods. This study confirms that FSL is superior to supervised and transfer learning in classifying metastatic cancer and demonstrates that using BSR as a loss function and modifying Finetune can enhance the model's capabilities.
Using Spatio-Temporal Dual-Stream Network with Self-Supervised Learning for Lung Tumor Classification on Radial Probe Endobronchial Ultrasound Video
May 07, 2023Abstract:The purpose of this study is to develop a computer-aided diagnosis system for classifying benign and malignant lung lesions, and to assist physicians in real-time analysis of radial probe endobronchial ultrasound (EBUS) videos. During the biopsy process of lung cancer, physicians use real-time ultrasound images to find suitable lesion locations for sampling. However, most of these images are difficult to classify and contain a lot of noise. Previous studies have employed 2D convolutional neural networks to effectively differentiate between benign and malignant lung lesions, but doctors still need to manually select good-quality images, which can result in additional labor costs. In addition, the 2D neural network has no ability to capture the temporal information of the ultrasound video, so it is difficult to obtain the relationship between the features of the continuous images. This study designs an automatic diagnosis system based on a 3D neural network, uses the SlowFast architecture as the backbone to fuse temporal and spatial features, and uses the SwAV method of contrastive learning to enhance the noise robustness of the model. The method we propose includes the following advantages, such as (1) using clinical ultrasound films as model input, thereby reducing the need for high-quality image selection by physicians, (2) high-accuracy classification of benign and malignant lung lesions can assist doctors in clinical diagnosis and reduce the time and risk of surgery, and (3) the capability to classify well even in the presence of significant image noise. The AUC, accuracy, precision, recall and specificity of our proposed method on the validation set reached 0.87, 83.87%, 86.96%, 90.91% and 66.67%, respectively. The results have verified the importance of incorporating temporal information and the effectiveness of using the method of contrastive learning on feature extraction.
The interpretation of endobronchial ultrasound image using 3D convolutional neural network for differentiating malignant and benign mediastinal lesions
Aug 02, 2021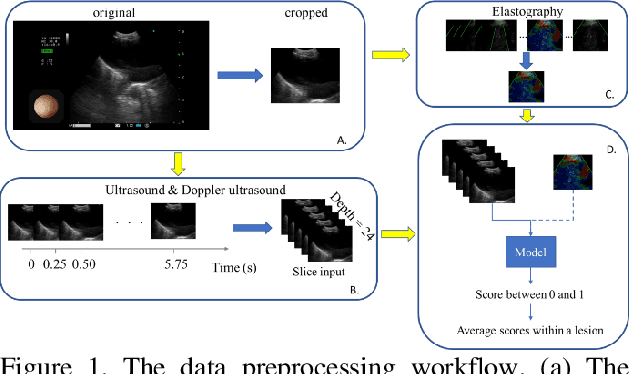
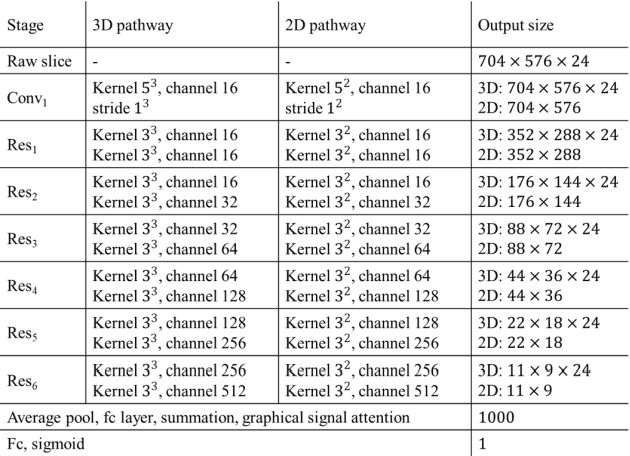
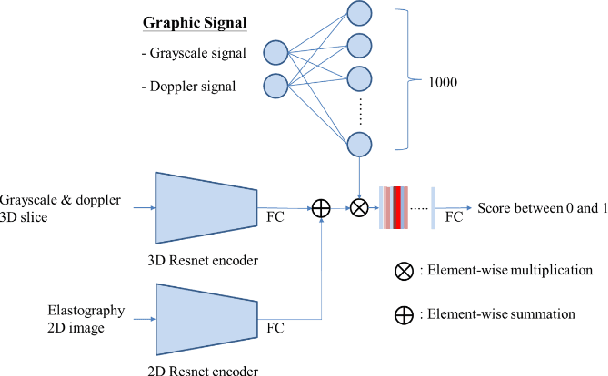
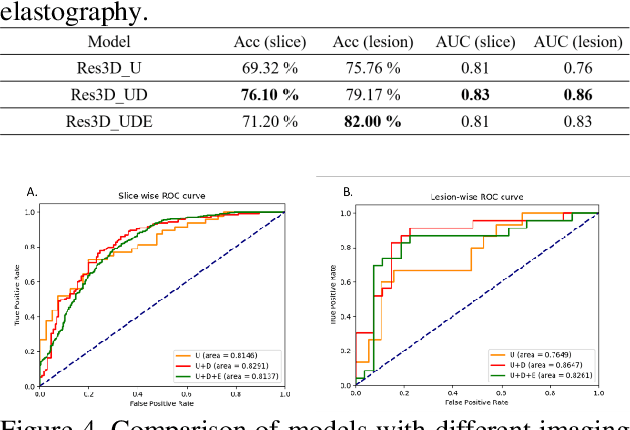
Abstract:The purpose of this study is to differentiate malignant and benign mediastinal lesions by using the three-dimensional convolutional neural network through the endobronchial ultrasound (EBUS) image. Compared with previous study, our proposed model is robust to noise and able to fuse various imaging features and spatiotemporal features of EBUS videos. Endobronchial ultrasound-guided transbronchial needle aspiration (EBUS-TBNA) is a diagnostic tool for intrathoracic lymph nodes. Physician can observe the characteristics of the lesion using grayscale mode, doppler mode, and elastography during the procedure. To process the EBUS data in the form of a video and appropriately integrate the features of multiple imaging modes, we used a time-series three-dimensional convolutional neural network (3D CNN) to learn the spatiotemporal features and design a variety of architectures to fuse each imaging mode. Our model (Res3D_UDE) took grayscale mode, Doppler mode, and elastography as training data and achieved an accuracy of 82.00% and area under the curve (AUC) of 0.83 on the validation set. Compared with previous study, we directly used videos recorded during procedure as training and validation data, without additional manual selection, which might be easier for clinical application. In addition, model designed with 3D CNN can also effectively learn spatiotemporal features and improve accuracy. In the future, our model may be used to guide physicians to quickly and correctly find the target lesions for slice sampling during the inspection process, reduce the number of slices of benign lesions, and shorten the inspection time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge