Jeonghyeon Kim
Enhanced OoD Detection through Cross-Modal Alignment of Multi-Modal Representations
Mar 24, 2025Abstract:Prior research on out-of-distribution detection (OoDD) has primarily focused on single-modality models. Recently, with the advent of large-scale pretrained vision-language models such as CLIP, OoDD methods utilizing such multi-modal representations through zero-shot and prompt learning strategies have emerged. However, these methods typically involve either freezing the pretrained weights or only partially tuning them, which can be suboptimal for downstream datasets. In this paper, we highlight that multi-modal fine-tuning (MMFT) can achieve notable OoDD performance. Despite some recent works demonstrating the impact of fine-tuning methods for OoDD, there remains significant potential for performance improvement. We investigate the limitation of na\"ive fine-tuning methods, examining why they fail to fully leverage the pretrained knowledge. Our empirical analysis suggests that this issue could stem from the modality gap within in-distribution (ID) embeddings. To address this, we propose a training objective that enhances cross-modal alignment by regularizing the distances between image and text embeddings of ID data. This adjustment helps in better utilizing pretrained textual information by aligning similar semantics from different modalities (i.e., text and image) more closely in the hyperspherical representation space. We theoretically demonstrate that the proposed regularization corresponds to the maximum likelihood estimation of an energy-based model on a hypersphere. Utilizing ImageNet-1k OoD benchmark datasets, we show that our method, combined with post-hoc OoDD approaches leveraging pretrained knowledge (e.g., NegLabel), significantly outperforms existing methods, achieving state-of-the-art OoDD performance and leading ID accuracy.
Understanding active learning of molecular docking and its applications
Jun 14, 2024Abstract:With the advancing capabilities of computational methodologies and resources, ultra-large-scale virtual screening via molecular docking has emerged as a prominent strategy for in silico hit discovery. Given the exhaustive nature of ultra-large-scale virtual screening, active learning methodologies have garnered attention as a means to mitigate computational cost through iterative small-scale docking and machine learning model training. While the efficacy of active learning methodologies has been empirically validated in extant literature, a critical investigation remains in how surrogate models can predict docking score without considering three-dimensional structural features, such as receptor conformation and binding poses. In this paper, we thus investigate how active learning methodologies effectively predict docking scores using only 2D structures and under what circumstances they may work particularly well through benchmark studies encompassing six receptor targets. Our findings suggest that surrogate models tend to memorize structural patterns prevalent in high docking scored compounds obtained during acquisition steps. Despite this tendency, surrogate models demonstrate utility in virtual screening, as exemplified in the identification of actives from DUD-E dataset and high docking-scored compounds from EnamineReal library, a significantly larger set than the initial screening pool. Our comprehensive analysis underscores the reliability and potential applicability of active learning methodologies in virtual screening campaigns.
Deep Active Learning with Contrastive Learning Under Realistic Data Pool Assumptions
Mar 25, 2023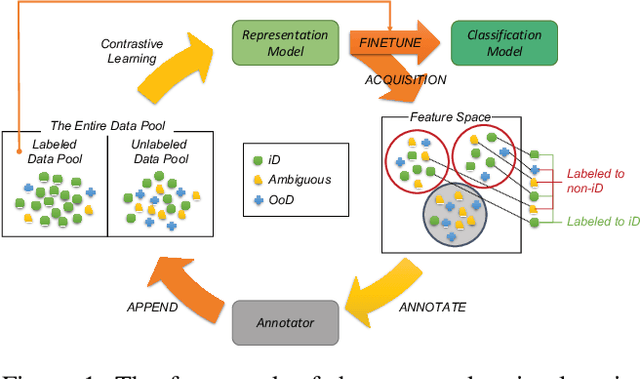
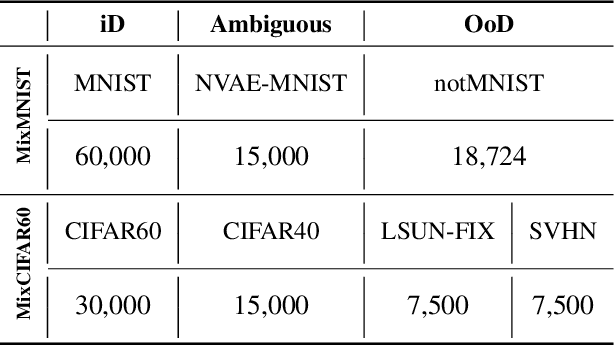
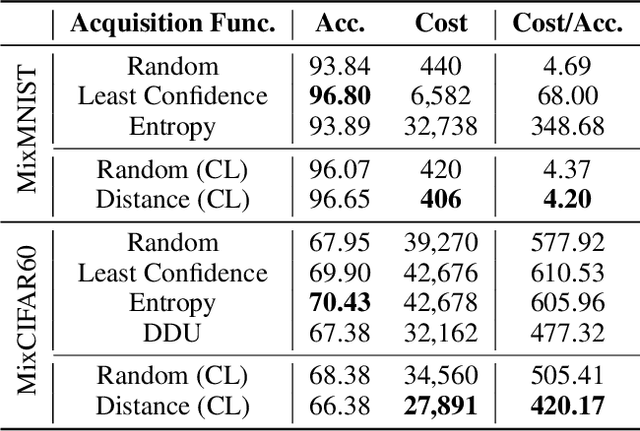
Abstract:Active learning aims to identify the most informative data from an unlabeled data pool that enables a model to reach the desired accuracy rapidly. This benefits especially deep neural networks which generally require a huge number of labeled samples to achieve high performance. Most existing active learning methods have been evaluated in an ideal setting where only samples relevant to the target task, i.e., in-distribution samples, exist in an unlabeled data pool. A data pool gathered from the wild, however, is likely to include samples that are irrelevant to the target task at all and/or too ambiguous to assign a single class label even for the oracle. We argue that assuming an unlabeled data pool consisting of samples from various distributions is more realistic. In this work, we introduce new active learning benchmarks that include ambiguous, task-irrelevant out-of-distribution as well as in-distribution samples. We also propose an active learning method designed to acquire informative in-distribution samples in priority. The proposed method leverages both labeled and unlabeled data pools and selects samples from clusters on the feature space constructed via contrastive learning. Experimental results demonstrate that the proposed method requires a lower annotation budget than existing active learning methods to reach the same level of accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge