Jeong Min Lee
Personalized Event Prediction for Electronic Health Records
Aug 21, 2023



Abstract:Clinical event sequences consist of hundreds of clinical events that represent records of patient care in time. Developing accurate predictive models of such sequences is of a great importance for supporting a variety of models for interpreting/classifying the current patient condition, or predicting adverse clinical events and outcomes, all aimed to improve patient care. One important challenge of learning predictive models of clinical sequences is their patient-specific variability. Based on underlying clinical conditions, each patient's sequence may consist of different sets of clinical events (observations, lab results, medications, procedures). Hence, simple population-wide models learned from event sequences for many different patients may not accurately predict patient-specific dynamics of event sequences and their differences. To address the problem, we propose and investigate multiple new event sequence prediction models and methods that let us better adjust the prediction for individual patients and their specific conditions. The methods developed in this work pursue refinement of population-wide models to subpopulations, self-adaptation, and a meta-level model switching that is able to adaptively select the model with the best chance to support the immediate prediction. We analyze and test the performance of these models on clinical event sequences of patients in MIMIC-III database.
* arXiv admin note: text overlap with arXiv:2104.01787
Learning to Adapt Clinical Sequences with Residual Mixture of Experts
Apr 06, 2022



Abstract:Clinical event sequences in Electronic Health Records (EHRs) record detailed information about the patient condition and patient care as they occur in time. Recent years have witnessed increased interest of machine learning community in developing machine learning models solving different types of problems defined upon information in EHRs. More recently, neural sequential models, such as RNN and LSTM, became popular and widely applied models for representing patient sequence data and for predicting future events or outcomes based on such data. However, a single neural sequential model may not properly represent complex dynamics of all patients and the differences in their behaviors. In this work, we aim to alleviate this limitation by refining a one-fits-all model using a Mixture-of-Experts (MoE) architecture. The architecture consists of multiple (expert) RNN models covering patient sub-populations and refining the predictions of the base model. That is, instead of training expert RNN models from scratch we define them on the residual signal that attempts to model the differences from the population-wide model. The heterogeneity of various patient sequences is modeled through multiple experts that consist of RNN. Particularly, instead of directly training MoE from scratch, we augment MoE based on the prediction signal from pretrained base GRU model. With this way, the mixture of experts can provide flexible adaptation to the (limited) predictive power of the single base RNN model. We experiment with the newly proposed model on real-world EHRs data and the multivariate clinical event prediction task. We implement RNN using Gated Recurrent Units (GRU). We show 4.1% gain on AUPRC statistics compared to a single RNN prediction.
Simultaneous super-resolution and motion artifact removal in diffusion-weighted MRI using unsupervised deep learning
May 01, 2021



Abstract:Diffusion-weighted MRI is nowadays performed routinely due to its prognostic ability, yet the quality of the scans are often unsatisfactory which can subsequently hamper the clinical utility. To overcome the limitations, here we propose a fully unsupervised quality enhancement scheme, which boosts the resolution and removes the motion artifact simultaneously. This process is done by first training the network using optimal transport driven cycleGAN with stochastic degradation block which learns to remove aliasing artifacts and enhance the resolution, then using the trained network in the test stage by utilizing bootstrap subsampling and aggregation for motion artifact suppression. We further show that we can control the trade-off between the amount of artifact correction and resolution by controlling the bootstrap subsampling ratio at the inference stage. To the best of our knowledge, the proposed method is the first to tackle super-resolution and motion artifact correction simultaneously in the context of MRI using unsupervised learning. We demonstrate the efficiency of our method by applying it to both quantitative evaluation using simulation study, and to in vivo diffusion-weighted MR scans, which shows that our method is superior to the current state-of-the-art methods. The proposed method is flexible in that it can be applied to various quality enhancement schemes in other types of MR scans, and also directly to the quality enhancement of apparent diffusion coefficient maps.
Neural Clinical Event Sequence Prediction through Personalized Online Adaptive Learning
Apr 06, 2021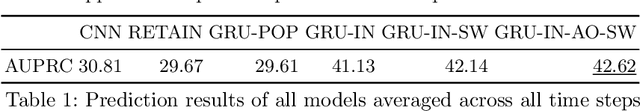
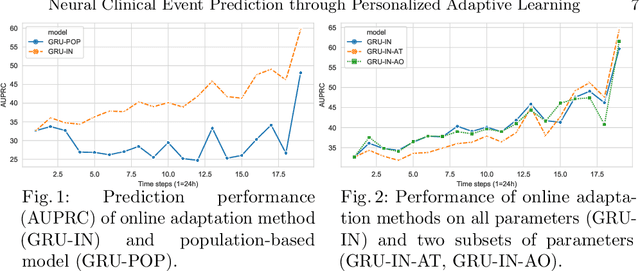
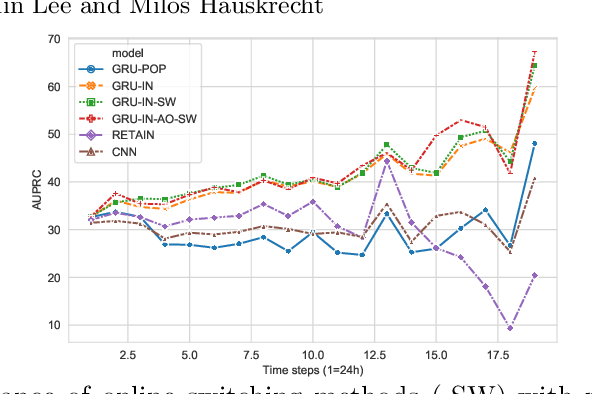
Abstract:Clinical event sequences consist of thousands of clinical events that represent records of patient care in time. Developing accurate prediction models for such sequences is of a great importance for defining representations of a patient state and for improving patient care. One important challenge of learning a good predictive model of clinical sequences is patient-specific variability. Based on underlying clinical complications, each patient's sequence may consist of different sets of clinical events. However, population-based models learned from such sequences may not accurately predict patient-specific dynamics of event sequences. To address the problem, we develop a new adaptive event sequence prediction framework that learns to adjust its prediction for individual patients through an online model update.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge