Jelica Vasiljević
HistoStarGAN: A Unified Approach to Stain Normalisation, Stain Transfer and Stain Invariant Segmentation in Renal Histopathology
Oct 18, 2022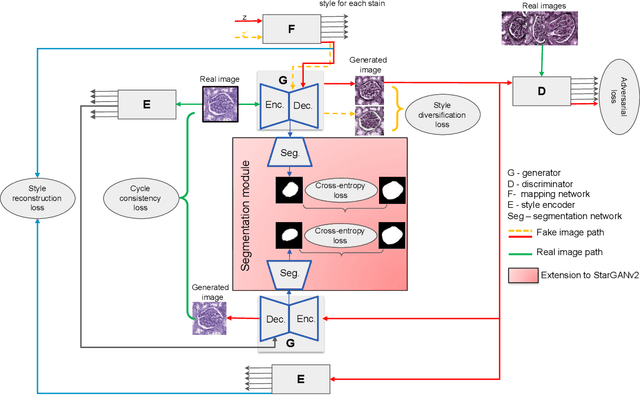

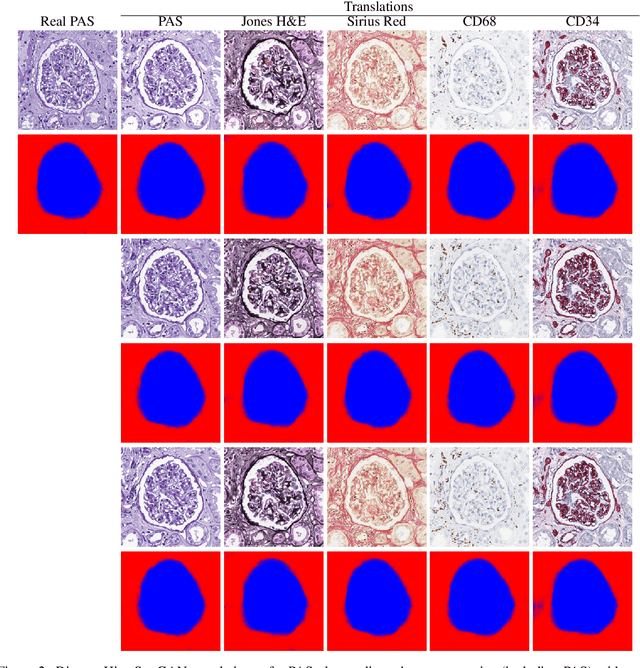
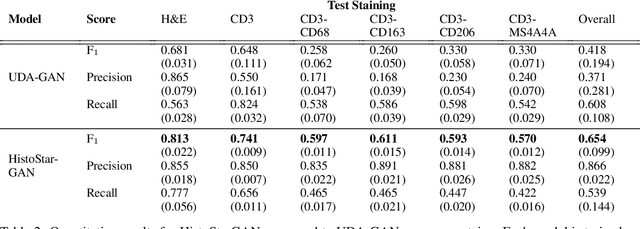
Abstract:Virtual stain transfer is a promising area of research in Computational Pathology, which has a great potential to alleviate important limitations when applying deeplearningbased solutions such as lack of annotations and sensitivity to a domain shift. However, in the literature, the majority of virtual staining approaches are trained for a specific staining or stain combination, and their extension to unseen stainings requires the acquisition of additional data and training. In this paper, we propose HistoStarGAN, a unified framework that performs stain transfer between multiple stainings, stain normalisation and stain invariant segmentation, all in one inference of the model. We demonstrate the generalisation abilities of the proposed solution to perform diverse stain transfer and accurate stain invariant segmentation over numerous unseen stainings, which is the first such demonstration in the field. Moreover, the pre-trained HistoStar-GAN model can serve as a synthetic data generator, which paves the way for the use of fully annotated synthetic image data to improve the training of deep learning-based algorithms. To illustrate the capabilities of our approach, as well as the potential risks in the microscopy domain, inspired by applications in natural images, we generated KidneyArtPathology, a fully annotated artificial image dataset for renal pathology.
Towards Measuring Domain Shift in Histopathological Stain Translation in an Unsupervised Manner
May 09, 2022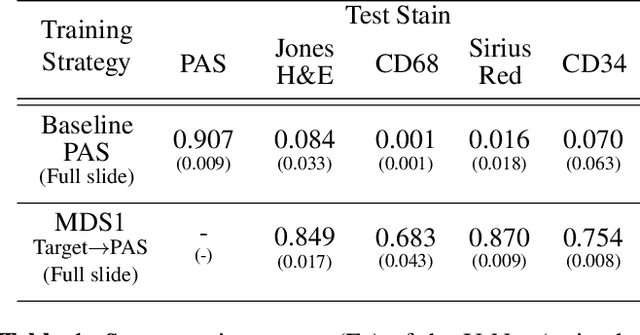
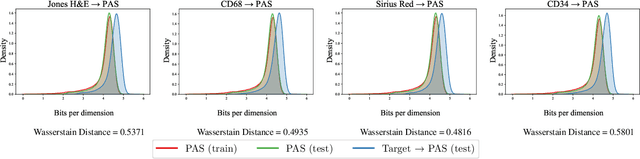
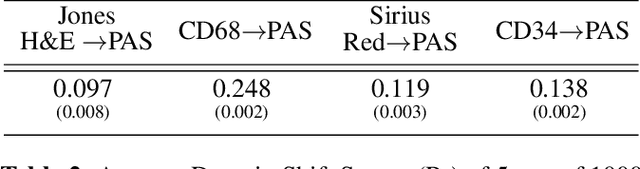
Abstract:Domain shift in digital histopathology can occur when different stains or scanners are used, during stain translation, etc. A deep neural network trained on source data may not generalise well to data that has undergone some domain shift. An important step towards being robust to domain shift is the ability to detect and measure it. This article demonstrates that the PixelCNN and domain shift metric can be used to detect and quantify domain shift in digital histopathology, and they demonstrate a strong correlation with generalisation performance. These findings pave the way for a mechanism to infer the average performance of a model (trained on source data) on unseen and unlabelled target data.
* 5 pages, 3 figures, 2 tables
Self adversarial attack as an augmentation method for immunohistochemical stainings
Mar 21, 2021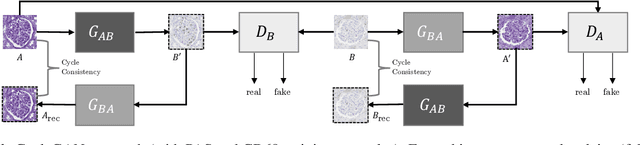
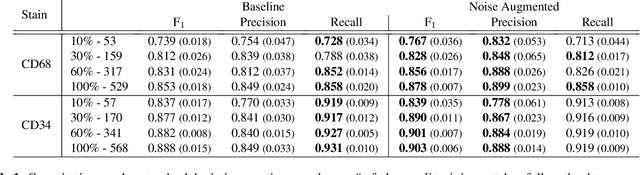

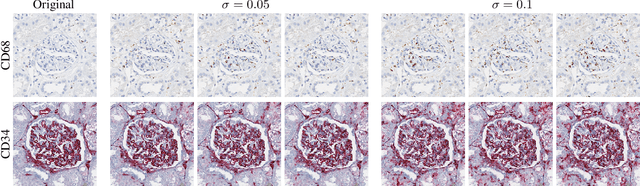
Abstract:It has been shown that unpaired image-to-image translation methods constrained by cycle-consistency hide the information necessary for accurate input reconstruction as imperceptible noise. We demonstrate that, when applied to histopathology data, this hidden noise appears to be related to stain specific features and show that this is the case with two immunohistochemical stainings during translation to Periodic acid- Schiff (PAS), a histochemical staining method commonly applied in renal pathology. Moreover, by perturbing this hidden information, the translation models produce different, plausible outputs. We demonstrate that this property can be used as an augmentation method which, in a case of supervised glomeruli segmentation, leads to improved performance.
Towards Histopathological Stain Invariance by Unsupervised Domain Augmentation using Generative Adversarial Networks
Dec 22, 2020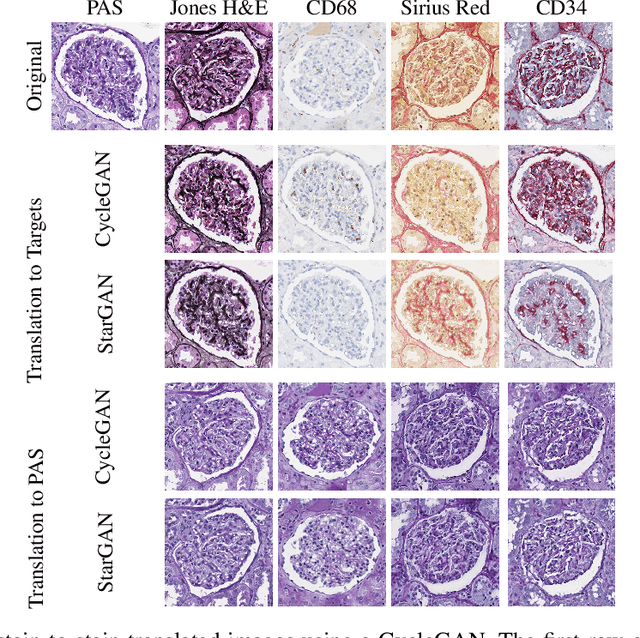
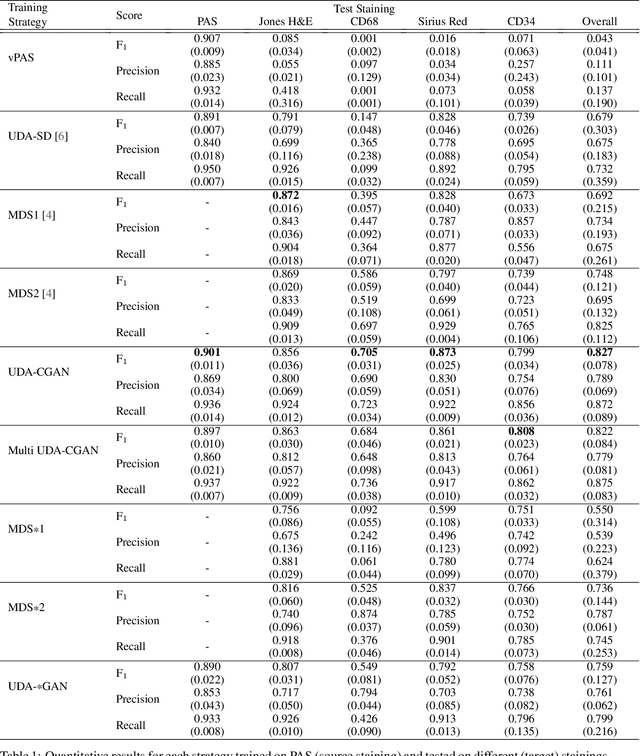
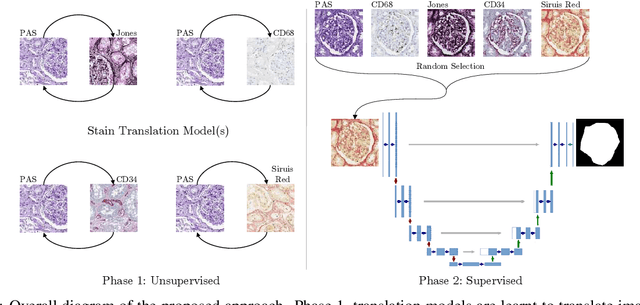
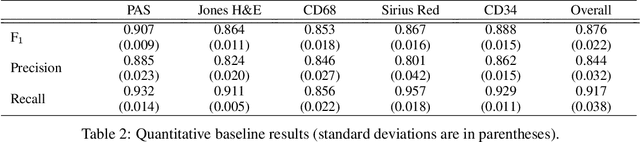
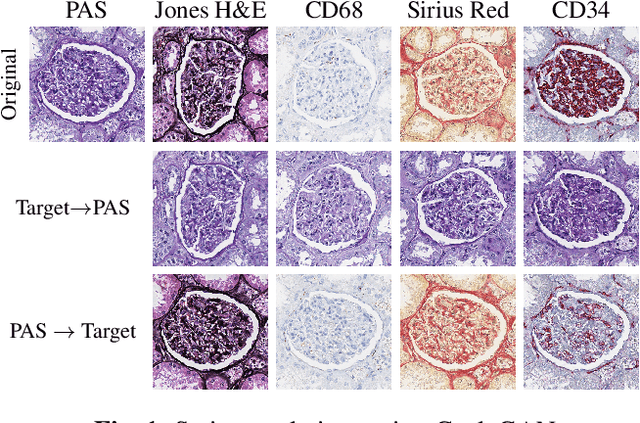
Abstract:The application of supervised deep learning methods in digital pathology is limited due to their sensitivity to domain shift. Digital Pathology is an area prone to high variability due to many sources, including the common practice of evaluating several consecutive tissue sections stained with different staining protocols. Obtaining labels for each stain is very expensive and time consuming as it requires a high level of domain knowledge. In this article, we propose an unsupervised augmentation approach based on adversarial image-to-image translation, which facilitates the training of stain invariant supervised convolutional neural networks. By training the network on one commonly used staining modality and applying it to images that include corresponding, but differently stained, tissue structures, the presented method demonstrates significant improvements over other approaches. These benefits are illustrated in the problem of glomeruli segmentation in seven different staining modalities (PAS, Jones H&E, CD68, Sirius Red, CD34, H&E and CD3) and analysis of the learned representations demonstrate their stain invariance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge