Jean-Baptiste Lamy
LIM\&BIO
A randomized simulation trial evaluating ABiMed, a clinical decision support system for medication reviews and polypharmacy management
Sep 03, 2024



Abstract:Background: Medication review is a structured interview of the patient, performed by the pharmacist and aimed at optimizing drug treatments. In practice, medication review is a long and cognitively-demanding task that requires specific knowledge. Clinical practice guidelines have been proposed, but their application is tedious. Methods: We designed ABiMed, a clinical decision support system for medication reviews, based on the implementation of the STOPP/START v2 guidelines and on the visual presentation of aggregated drug knowledge using tables, graphs and flower glyphs. We evaluated ABiMed with 39 community pharmacists during a randomized simulation trial, each pharmacist performing a medication review for two fictitious patients without ABiMed, and two others with ABiMed. We recorded the problems identified by the pharmacists, the interventions proposed, the response time, the perceived usability and the comments. Pharmacists' medication reviews were compared to an expert-designed gold standard. Results: With ABiMed, pharmacists found 1.6 times more relevant drug-related problems during the medication review (p=1.1e-12) and proposed better interventions (p=9.8e-9), without needing more time (p=0.56). The System Usability Scale score is 82.7, which is ranked "excellent". In their comments, pharmacists appreciated the visual aspect of ABiMed and its ability to compare the current treatment with the proposed one. A multifactor analysis showed no difference in the support offered by ABiMed according to the pharmacist's age or sex, in terms of percentage of problems identified or quality of the proposed interventions. Conclusions: The use of an intelligent and visual clinical decision support system can help pharmacists when they perform medication reviews. Our main perspective is the validation of the system in clinical conditions.
ABiMed: An intelligent and visual clinical decision support system for medication reviews and polypharmacy management
Dec 13, 2023Abstract:Background: Polypharmacy, i.e. taking five drugs or more, is both a public health and an economic issue. Medication reviews are structured interviews of the patient by the community pharmacist, aiming at optimizing the drug treatment and deprescribing useless, redundant or dangerous drugs. However, they remain difficult to perform and time-consuming. Several clinical decision support systems were developed for helping clinicians to manage polypharmacy. However, most were limited to the implementation of clinical practice guidelines. In this work, our objective is to design an innovative clinical decision support system for medication reviews and polypharmacy management, named ABiMed. Methods: ABiMed associates several approaches: guidelines implementation, but the automatic extraction of patient data from the GP's electronic health record and its transfer to the pharmacist, and the visual presentation of contextualized drug knowledge using visual analytics. We performed an ergonomic assessment and qualitative evaluations involving pharmacists and GPs during focus groups and workshops. Results: We describe the proposed architecture, which allows a collaborative multi-user usage. We present the various screens of ABiMed for entering or verifying patient data, for accessing drug knowledge (posology, adverse effects, interactions), for viewing STOPP/START rules and for suggesting modification to the treatment. Qualitative evaluations showed that health professionals were highly interested by our approach, associating the automatic guidelines execution with the visual presentation of drug knowledge. Conclusions: The association of guidelines implementation with visual presentation of knowledge is a promising approach for managing polypharmacy. Future works will focus on the improvement and the evaluation of ABiMed.
Adaptive questionnaires for facilitating patient data entry in clinical decision support systems: Methods and application to STOPP/START v2
Sep 19, 2023


Abstract:Clinical decision support systems are software tools that help clinicians to make medical decisions. However, their acceptance by clinicians is usually rather low. A known problem is that they often require clinicians to manually enter lots of patient data, which is long and tedious. Existing solutions, such as the automatic data extraction from electronic health record, are not fully satisfying, because of low data quality and availability. In practice, many systems still include long questionnaire for data entry. In this paper, we propose an original solution to simplify patient data entry, using an adaptive questionnaire, i.e. a questionnaire that evolves during user interaction, showing or hiding questions dynamically. Considering a rule-based decision support systems, we designed methods for translating the system's clinical rules into display rules that determine the items to show in the questionnaire, and methods for determining the optimal order of priority among the items in the questionnaire. We applied this approach to a decision support system implementing STOPP/START v2, a guideline for managing polypharmacy. We show that it permits reducing by about two thirds the number of clinical conditions displayed in the questionnaire. Presented to clinicians during focus group sessions, the adaptive questionnaire was found "pretty easy to use". In the future, this approach could be applied to other guidelines, and adapted for data entry by patients.
A data science approach to drug safety: Semantic and visual mining of adverse drug events from clinical trials of pain treatments
Jun 19, 2020



Abstract:Clinical trials are the basis of Evidence-Based Medicine. Trial results are reviewed by experts and consensus panels for producing meta-analyses and clinical practice guidelines. However, reviewing these results is a long and tedious task, hence the meta-analyses and guidelines are not updated each time a new trial is published. Moreover, the independence of experts may be difficult to appraise. On the contrary, in many other domains, including medical risk analysis, the advent of data science, big data and visual analytics allowed moving from expert-based to fact-based knowledge. Since 12 years, many trial results are publicly available online in trial registries. Nevertheless, data science methods have not yet been applied widely to trial data. In this paper, we present a platform for analyzing the safety events reported during clinical trials and published in trial registries. This platform is based on an ontological model including 582 trials on pain treatments, and uses semantic web technologies for querying this dataset at various levels of granularity. It also relies on a 26-dimensional flower glyph for the visualization of the Adverse Drug Events (ADE) rates in 13 categories and 2 levels of seriousness. We illustrate the interest of this platform through several use cases and we were able to find back conclusions that are known in the literature. The platform was presented to four experts in drug safety, and is publicly available online, with the ontology of pain treatment ADE.
Using graph transformation algorithms to generate natural language equivalents of icons expressing medical concepts
Sep 26, 2014
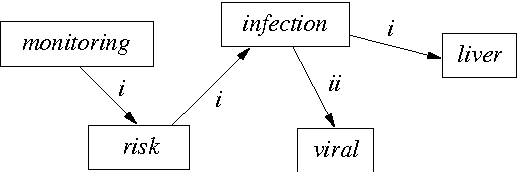
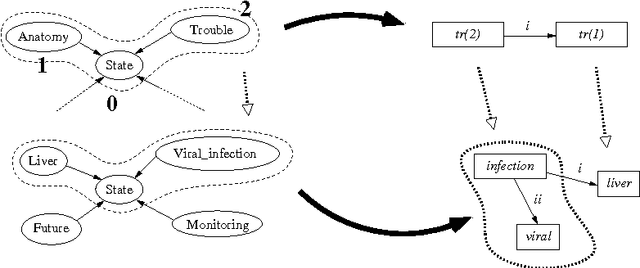
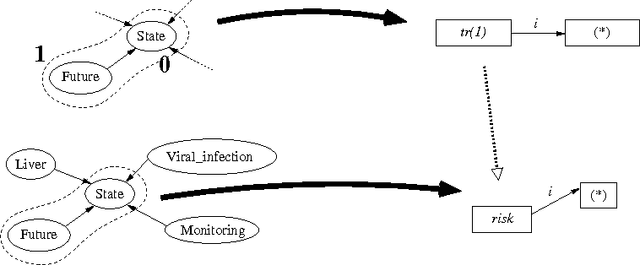
Abstract:A graphical language addresses the need to communicate medical information in a synthetic way. Medical concepts are expressed by icons conveying fast visual information about patients' current state or about the known effects of drugs. In order to increase the visual language's acceptance and usability, a natural language generation interface is currently developed. In this context, this paper describes the use of an informatics method ---graph transformation--- to prepare data consisting of concepts in an OWL-DL ontology for use in a natural language generation component. The OWL concept may be considered as a star-shaped graph with a central node. The method transforms it into a graph representing the deep semantic structure of a natural language phrase. This work may be of future use in other contexts where ontology concepts have to be mapped to half-formalized natural language expressions.
A semi-automatic semantic method for mapping SNOMED CT concepts to VCM Icons
Dec 03, 2013

Abstract:VCM (Visualization of Concept in Medicine) is an iconic language for representing key medical concepts by icons. However, the use of this language with reference terminologies, such as SNOMED CT, will require the mapping of its icons to the terms of these terminologies. Here, we present and evaluate a semi-automatic semantic method for the mapping of SNOMED CT concepts to VCM icons. Both SNOMED CT and VCM are compositional in nature; SNOMED CT is expressed in description logic and VCM semantics are formalized in an OWL ontology. The proposed method involves the manual mapping of a limited number of underlying concepts from the VCM ontology, followed by automatic generation of the rest of the mapping. We applied this method to the clinical findings of the SNOMED CT CORE subset, and 100 randomly-selected mappings were evaluated by three experts. The results obtained were promising, with 82 of the SNOMED CT concepts correctly linked to VCM icons according to the experts. Most of the errors were easy to fix.
A generic system for critiquing physicians' prescriptions: usability, satisfaction and lessons learnt
Dec 03, 2013
Abstract:Clinical decision support systems have been developed to help physicians to take clinical guidelines into account during consultations. The ASTI critiquing module is one such systems; it provides the physician with automatic criticisms when a drug prescription does not follow the guidelines. It was initially developed for hypertension and type 2 diabetes, but is designed to be generic enough for application to all chronic diseases. We present here the results of usability and satisfaction evaluations for the ASTI critiquing module, obtained with GPs for a newly implemented guideline concerning dyslipaemia, and we discuss the lessons learnt and the difficulties encountered when building a generic DSS for critiquing physicians' prescriptions.
Use of the C4.5 machine learning algorithm to test a clinical guideline-based decision support system
Dec 03, 2013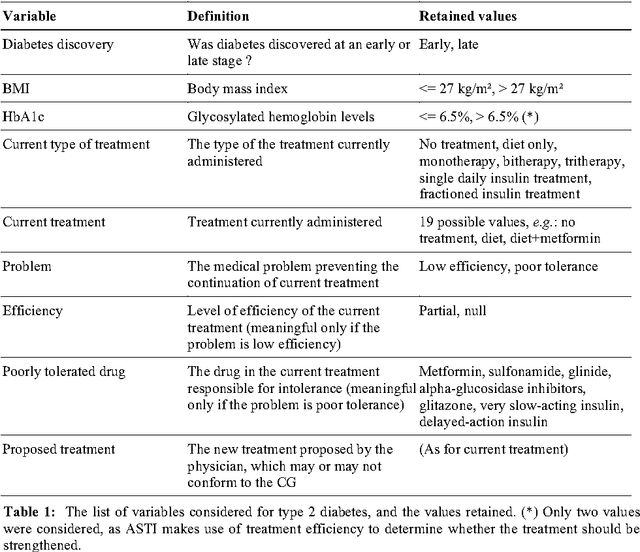

Abstract:Well-designed medical decision support system (DSS) have been shown to improve health care quality. However, before they can be used in real clinical situations, these systems must be extensively tested, to ensure that they conform to the clinical guidelines (CG) on which they are based. Existing methods cannot be used for the systematic testing of all possible test cases. We describe here a new exhaustive dynamic verification method. In this method, the DSS is considered to be a black box, and the Quinlan C4.5 algorithm is used to build a decision tree from an exhaustive set of DSS input vectors and outputs. This method was successfully used for the testing of a medical DSS relating to chronic diseases: the ASTI critiquing module for type 2 diabetes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge