Jason Holmberg
The SA-FARI Dataset: Segment Anything in Footage of Animals for Recognition and Identification
Nov 19, 2025Abstract:Automated video analysis is critical for wildlife conservation. A foundational task in this domain is multi-animal tracking (MAT), which underpins applications such as individual re-identification and behavior recognition. However, existing datasets are limited in scale, constrained to a few species, or lack sufficient temporal and geographical diversity - leaving no suitable benchmark for training general-purpose MAT models applicable across wild animal populations. To address this, we introduce SA-FARI, the largest open-source MAT dataset for wild animals. It comprises 11,609 camera trap videos collected over approximately 10 years (2014-2024) from 741 locations across 4 continents, spanning 99 species categories. Each video is exhaustively annotated culminating in ~46 hours of densely annotated footage containing 16,224 masklet identities and 942,702 individual bounding boxes, segmentation masks, and species labels. Alongside the task-specific annotations, we publish anonymized camera trap locations for each video. Finally, we present comprehensive benchmarks on SA-FARI using state-of-the-art vision-language models for detection and tracking, including SAM 3, evaluated with both species-specific and generic animal prompts. We also compare against vision-only methods developed specifically for wildlife analysis. SA-FARI is the first large-scale dataset to combine high species diversity, multi-region coverage, and high-quality spatio-temporal annotations, offering a new foundation for advancing generalizable multianimal tracking in the wild. The dataset is available at $\href{https://www.conservationxlabs.com/sa-fari}{\text{conservationxlabs.com/SA-FARI}}$.
Multispecies Animal Re-ID Using a Large Community-Curated Dataset
Dec 07, 2024Abstract:Recent work has established the ecological importance of developing algorithms for identifying animals individually from images. Typically, a separate algorithm is trained for each species, a natural step but one that creates significant barriers to wide-spread use: (1) each effort is expensive, requiring data collection, data curation, and model training, deployment, and maintenance, (2) there is little training data for many species, and (3) commonalities in appearance across species are not exploited. We propose an alternative approach focused on training multi-species individual identification (re-id) models. We construct a dataset that includes 49 species, 37K individual animals, and 225K images, using this data to train a single embedding network for all species. Our model employs an EfficientNetV2 backbone and a sub-center ArcFace loss function with dynamic margins. We evaluate the performance of this multispecies model in several ways. Most notably, we demonstrate that it consistently outperforms models trained separately on each species, achieving an average gain of 12.5% in top-1 accuracy. Furthermore, the model demonstrates strong zero-shot performance and fine-tuning capabilities for new species with limited training data, enabling effective curation of new species through both incremental addition of data to the training set and fine-tuning without the original data. Additionally, our model surpasses the recent MegaDescriptor on unseen species, averaging an 19.2% top-1 improvement per species and showing gains across all 33 species tested. The fully-featured code repository is publicly available on GitHub, and the feature extractor model can be accessed on HuggingFace for seamless integration with wildlife re-identification pipelines. The model is already in production use for 60+ species in a large-scale wildlife monitoring system.
Adapting the re-ID challenge for static sensors
Nov 30, 2024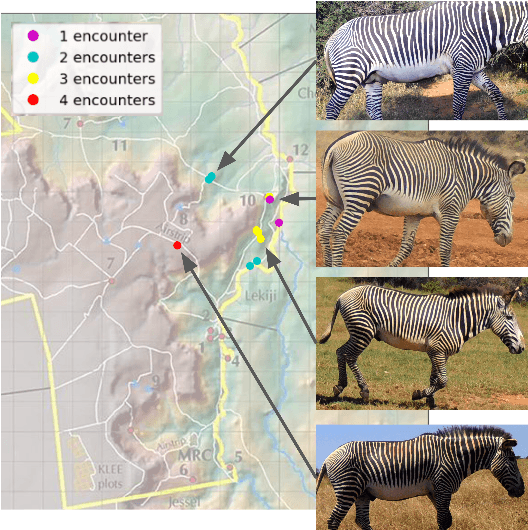
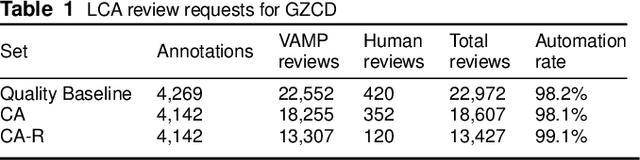
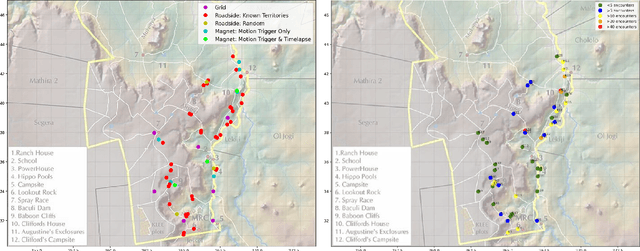
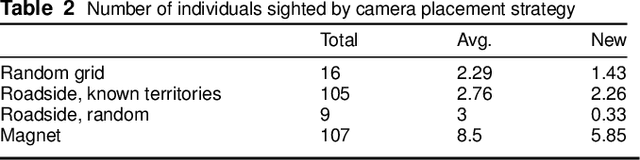
Abstract:In both 2016 and 2018, a census of the highly-endangered Grevy's zebra population was enabled by the Great Grevy's Rally (GGR), a citizen science event that produces population estimates via expert and algorithmic curation of volunteer-captured images. A complementary, scalable, and long-term Grevy's population monitoring approach involves deploying camera trap networks. However, in both scenarios, a substantial majority of zebra images are not usable for individual identification due to poor in-the-wild imaging conditions; camera trap images in particular present high rates of occlusion and high spatio-temporal similarity within image bursts. Our proposed filtering pipeline incorporates animal detection, species identification, viewpoint estimation, quality evaluation, and temporal subsampling to obtain individual crops suitable for re-ID, which are subsequently curated by the LCA decision management algorithm. Our method processed images taken during GGR-16 and GGR-18 in Meru County, Kenya, into 4,142 highly-comparable annotations, requiring only 120 contrastive human decisions to produce a population estimate within 4.6% of the ground-truth count. Our method also efficiently processed 8.9M unlabeled camera trap images from 70 cameras at the Mpala Research Centre in Laikipia County, Kenya over two years into 685 encounters of 173 individuals, requiring only 331 contrastive human decisions.
The Animal ID Problem: Continual Curation
Jun 18, 2021
Abstract:Hoping to stimulate new research in individual animal identification from images, we propose to formulate the problem as the human-machine Continual Curation of images and animal identities. This is an open world recognition problem, where most new animals enter the system after its algorithms are initially trained and deployed. Continual Curation, as defined here, requires (1) an improvement in the effectiveness of current recognition methods, (2) a pairwise verification algorithm that allows the possibility of no decision, and (3) an algorithmic decision mechanism that seeks human input to guide the curation process. Error metrics must evaluate the ability of recognition algorithms to identify not only animals that have been seen just once or twice but also recognize new animals not in the database. An important measure of overall system performance is accuracy as a function of the amount of human input required.
Integral Curvature Representation and Matching Algorithms for Identification of Dolphins and Whales
Aug 25, 2017

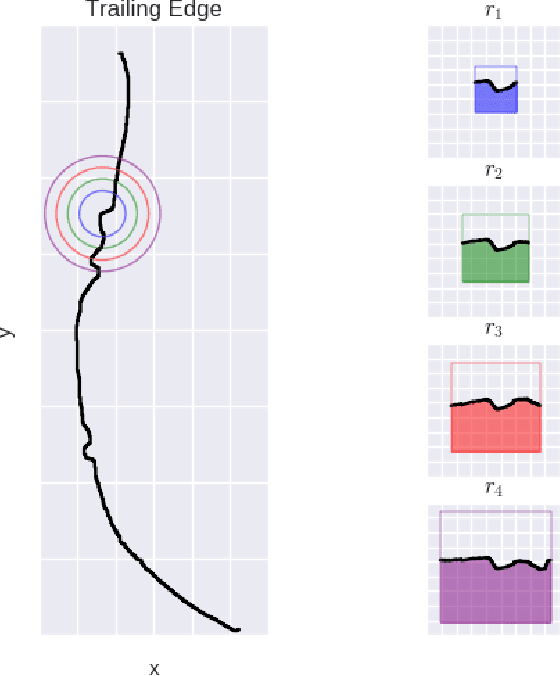
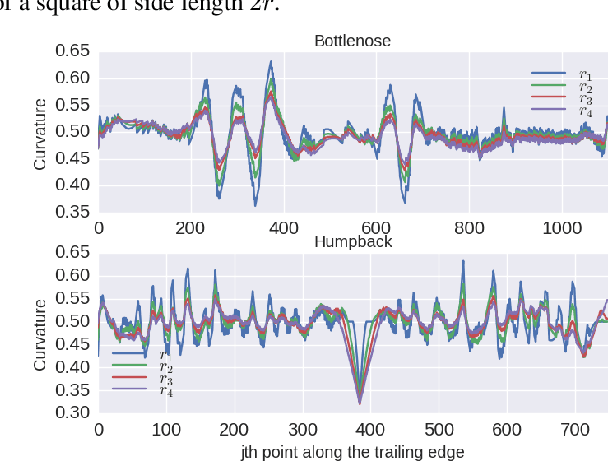
Abstract:We address the problem of identifying individual cetaceans from images showing the trailing edge of their fins. Given the trailing edge from an unknown individual, we produce a ranking of known individuals from a database. The nicks and notches along the trailing edge define an individual's unique signature. We define a representation based on integral curvature that is robust to changes in viewpoint and pose, and captures the pattern of nicks and notches in a local neighborhood at multiple scales. We explore two ranking methods that use this representation. The first uses a dynamic programming time-warping algorithm to align two representations, and interprets the alignment cost as a measure of similarity. This algorithm also exploits learned spatial weights to downweight matches from regions of unstable curvature. The second interprets the representation as a feature descriptor. Feature keypoints are defined at the local extrema of the representation. Descriptors for the set of known individuals are stored in a tree structure, which allows us to perform queries given the descriptors from an unknown trailing edge. We evaluate the top-k accuracy on two real-world datasets to demonstrate the effectiveness of the curvature representation, achieving top-1 accuracy scores of approximately 95% and 80% for bottlenose dolphins and humpback whales, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge