Janka Hatvani
pyPCG: A Python Toolbox Specialized for Phonocardiography Analysis
Aug 23, 2024Abstract:Phonocardiography has recently gained popularity in low-cost and remote monitoring, including passive fetal heart monitoring. Development for methods which analyse phonocardiographical data try to capitalize on this opportunity, and in recent years a multitude of such algorithms and models have been published. Although there is little to no standardization in these published algorithms and multiple parts of these models have to be reimplemented on a case-by-case basis. Datasets containing heart sound recordings also lack standardization in both data storage and labeling, especially in fetal phonocardiography. We are presenting a toolbox that can serve as a basis for a future standard framework for heart sound analysis. This toolbox contains some of the most widely used processing steps, and with these, complex analysis processes can be created. These functions can be individually tested. Due to the interdependence of the steps, we validated the current segmentation stage using a manually labeled fetal phonocardiogram dataset comprising 50 one-minute abdominal PCG recordings, which include 6,758 S1 and 6,729 S2 labels. Our results were compared to other common and available segmentation methods, peak detection with the Neurokit2 library, and the Hidden Semi-Markov Model by Springer et al. With a 30 ms tolerance our best model achieved a 97.1% F1 score and 10.8 +/- 7.9 ms mean absolute error for S1 detection. This detection accuracy outperformed all tested methods. With this a more accurate S2 detection method can be created as a multi-step process. After an accurate segmentation the extracted features should be representative of the selected segments, which allows for more accurate statistics or classification models. The toolbox contains functions for both feature extraction and statistics creation which are compatible with the previous steps.
A Tensor Factorization Method for 3D Super-Resolution with Application to Dental CT
Jul 26, 2018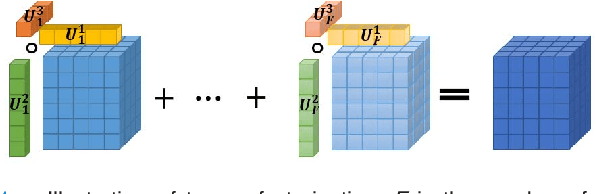
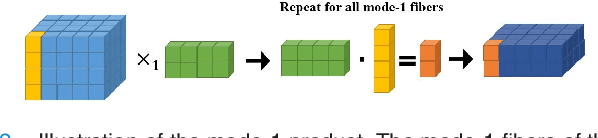
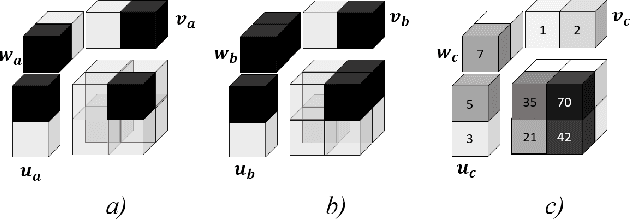
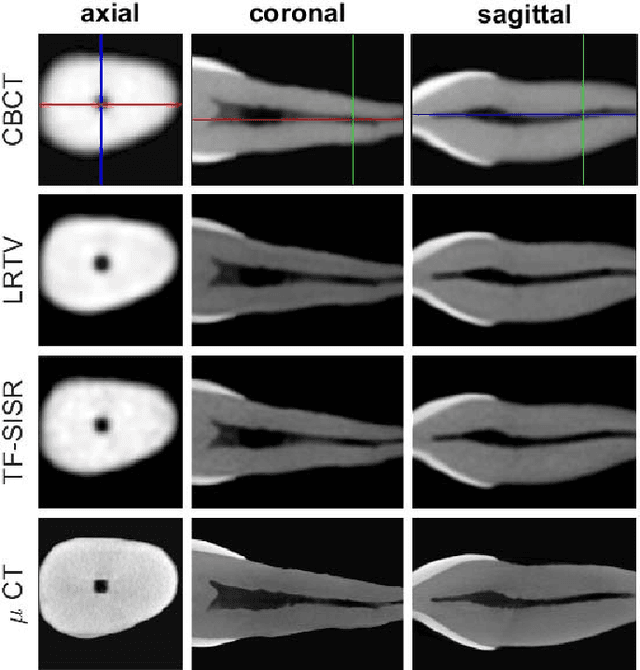
Abstract:Available super-resolution techniques for 3D images are either computationally inefficient prior-knowledge-based iterative techniques or deep learning methods which require a large database of known low- and high-resolution image pairs. A recently introduced tensor-factorization-based approach offers a fast solution without the use of known image pairs or strict prior assumptions. In this article this factorization framework is investigated for single image resolution enhancement with an off-line estimate of the system point spread function. The technique is applied to 3D cone beam computed tomography for dental image resolution enhancement. To demonstrate the efficiency of our method, it is compared to a recent state-of-the-art iterative technique using low-rank and total variation regularizations. In contrast to this comparative technique, the proposed reconstruction technique gives a 2-order-of-magnitude improvement in running time -- 2 minutes compared to 2 hours for a dental volume of 282$\times$266$\times$392 voxels. Furthermore, it also offers slightly improved quantitative results (peak signal-to-noise ratio, segmentation quality). Another advantage of the presented technique is the low number of hyperparameters. As demonstrated in this paper, the framework is not sensitive to small changes of its parameters, proposing an ease of use.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge