Irene Kim
Understanding Deep Neural Networks through Input Uncertainties
Nov 01, 2018

Abstract:Techniques for understanding the functioning of complex machine learning models are becoming increasingly popular, not only to improve the validation process, but also to extract new insights about the data via exploratory analysis. Though a large class of such tools currently exists, most assume that predictions are point estimates and use a sensitivity analysis of these estimates to interpret the model. Using lightweight probabilistic networks we show how including prediction uncertainties in the sensitivity analysis leads to: (i) more robust and generalizable models; and (ii) a new approach for model interpretation through uncertainty decomposition. In particular, we introduce a new regularization that takes both the mean and variance of a prediction into account and demonstrate that the resulting networks provide improved generalization to unseen data. Furthermore, we propose a new technique to explain prediction uncertainties through uncertainties in the input domain, thus providing new ways to validate and interpret deep learning models.
Autism Spectrum Disorder Classification using Graph Kernels on Multidimensional Time Series
Nov 29, 2016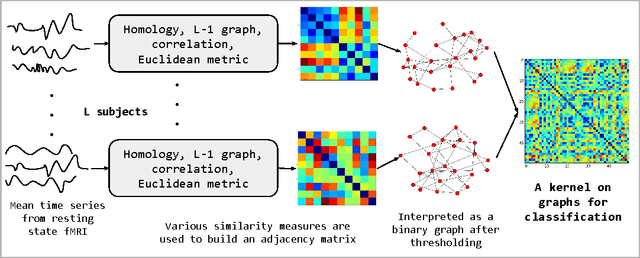

Abstract:We present an approach to model time series data from resting state fMRI for autism spectrum disorder (ASD) severity classification. We propose to adopt kernel machines and employ graph kernels that define a kernel dot product between two graphs. This enables us to take advantage of spatio-temporal information to capture the dynamics of the brain network, as opposed to aggregating them in the spatial or temporal dimension. In addition to the conventional similarity graphs, we explore the use of L1 graph using sparse coding, and the persistent homology of time delay embeddings, in the proposed pipeline for ASD classification. In our experiments on two datasets from the ABIDE collection, we demonstrate a consistent and significant advantage in using graph kernels over traditional linear or non linear kernels for a variety of time series features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge