Intekhab Hossain
Not all tickets are equal and we know it: Guiding pruning with domain-specific knowledge
Mar 05, 2024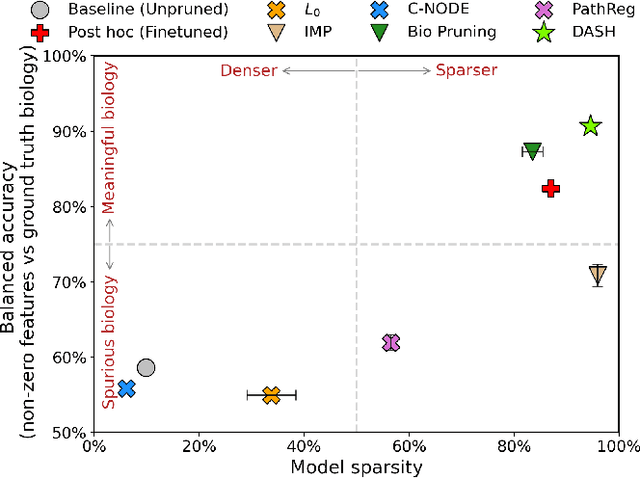

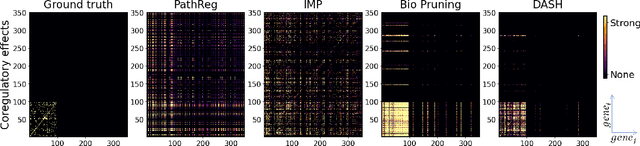

Abstract:Neural structure learning is of paramount importance for scientific discovery and interpretability. Yet, contemporary pruning algorithms that focus on computational resource efficiency face algorithmic barriers to select a meaningful model that aligns with domain expertise. To mitigate this challenge, we propose DASH, which guides pruning by available domain-specific structural information. In the context of learning dynamic gene regulatory network models, we show that DASH combined with existing general knowledge on interaction partners provides data-specific insights aligned with biology. For this task, we show on synthetic data with ground truth information and two real world applications the effectiveness of DASH, which outperforms competing methods by a large margin and provides more meaningful biological insights. Our work shows that domain specific structural information bears the potential to improve model-derived scientific insights.
A distribution-free mixed-integer optimization approach to hierarchical modelling of clustered and longitudinal data
Feb 06, 2023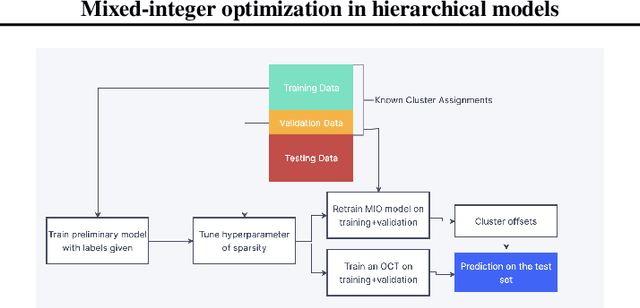



Abstract:We create a mixed-integer optimization (MIO) approach for doing cluster-aware regression, i.e. linear regression that takes into account the inherent clustered structure of the data. We compare to the linear mixed effects regression (LMEM) which is the most used current method, and design simulation experiments to show superior performance to LMEM in terms of both predictive and inferential metrics in silico. Furthermore, we show how our method is formulated in a very interpretable way; LMEM cannot generalize and make cluster-informed predictions when the cluster of new data points is unknown, but we solve this problem by training an interpretable classification tree that can help decide cluster effects for new data points, and demonstrate the power of this generalizability on a real protein expression dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge