Ilya Tyagin
QAOA-GPT: Efficient Generation of Adaptive and Regular Quantum Approximate Optimization Algorithm Circuits
Apr 23, 2025Abstract:Quantum computing has the potential to improve our ability to solve certain optimization problems that are computationally difficult for classical computers, by offering new algorithmic approaches that may provide speedups under specific conditions. In this work, we introduce QAOA-GPT, a generative framework that leverages Generative Pretrained Transformers (GPT) to directly synthesize quantum circuits for solving quadratic unconstrained binary optimization problems, and demonstrate it on the MaxCut problem on graphs. To diversify the training circuits and ensure their quality, we have generated a synthetic dataset using the adaptive QAOA approach, a method that incrementally builds and optimizes problem-specific circuits. The experiments conducted on a curated set of graph instances demonstrate that QAOA-GPT, generates high quality quantum circuits for new problem instances unseen in the training as well as successfully parametrizes QAOA. Our results show that using QAOA-GPT to generate quantum circuits will significantly decrease both the computational overhead of classical QAOA and adaptive approaches that often use gradient evaluation to generate the circuit and the classical optimization of the circuit parameters. Our work shows that generative AI could be a promising avenue to generate compact quantum circuits in a scalable way.
Dyport: Dynamic Importance-based Hypothesis Generation Benchmarking Technique
Dec 06, 2023Abstract:This paper presents a novel benchmarking framework Dyport for evaluating biomedical hypothesis generation systems. Utilizing curated datasets, our approach tests these systems under realistic conditions, enhancing the relevance of our evaluations. We integrate knowledge from the curated databases into a dynamic graph, accompanied by a method to quantify discovery importance. This not only assesses hypothesis accuracy but also their potential impact in biomedical research which significantly extends traditional link prediction benchmarks. Applicability of our benchmarking process is demonstrated on several link prediction systems applied on biomedical semantic knowledge graphs. Being flexible, our benchmarking system is designed for broad application in hypothesis generation quality verification, aiming to expand the scope of scientific discovery within the biomedical research community. Availability and implementation: Dyport framework is fully open-source. All code and datasets are available at: https://github.com/IlyaTyagin/Dyport
Literature-based Discovery for Landscape Planning
Jun 05, 2023Abstract:This project demonstrates how medical corpus hypothesis generation, a knowledge discovery field of AI, can be used to derive new research angles for landscape and urban planners. The hypothesis generation approach herein consists of a combination of deep learning with topic modeling, a probabilistic approach to natural language analysis that scans aggregated research databases for words that can be grouped together based on their subject matter commonalities; the word groups accordingly form topics that can provide implicit connections between two general research terms. The hypothesis generation system AGATHA was used to identify likely conceptual relationships between emerging infectious diseases (EIDs) and deforestation, with the objective of providing landscape planners guidelines for productive research directions to help them formulate research hypotheses centered on deforestation and EIDs that will contribute to the broader health field that asserts causal roles of landscape-level issues. This research also serves as a partial proof-of-concept for the application of medical database hypothesis generation to medicine-adjacent hypothesis discovery.
Towards Practical Explainability with Cluster Descriptors
Oct 20, 2022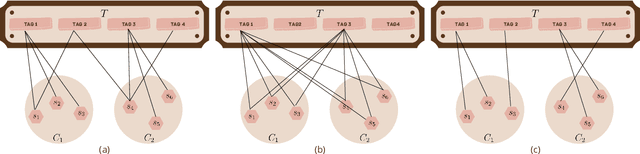

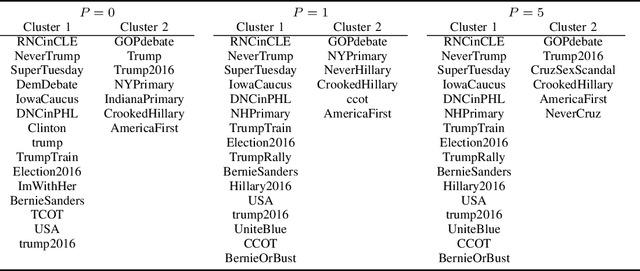
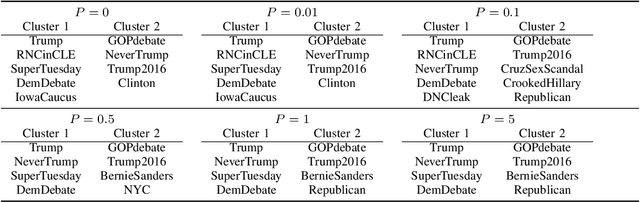
Abstract:With the rapid development of machine learning, improving its explainability has become a crucial research goal. We study the problem of making the clusters more explainable by investigating the cluster descriptors. Given a set of objects $S$, a clustering of these objects $\pi$, and a set of tags $T$ that have not participated in the clustering algorithm. Each object in $S$ is associated with a subset of $T$. The goal is to find a representative set of tags for each cluster, referred to as the cluster descriptors, with the constraint that these descriptors we find are pairwise disjoint, and the total size of all the descriptors is minimized. In general, this problem is NP-hard. We propose a novel explainability model that reinforces the previous models in such a way that tags that do not contribute to explainability and do not sufficiently distinguish between clusters are not added to the optimal descriptors. The proposed model is formulated as a quadratic unconstrained binary optimization problem which makes it suitable for solving on modern optimization hardware accelerators. We experimentally demonstrate how a proposed explainability model can be solved on specialized hardware for accelerating combinatorial optimization, the Fujitsu Digital Annealer, and use real-life Twitter and PubMed datasets for use cases.
Accelerating COVID-19 research with graph mining and transformer-based learning
Feb 10, 2021Abstract:In 2020, the White House released the, "Call to Action to the Tech Community on New Machine Readable COVID-19 Dataset," wherein artificial intelligence experts are asked to collect data and develop text mining techniques that can help the science community answer high-priority scientific questions related to COVID-19. The Allen Institute for AI and collaborators announced the availability of a rapidly growing open dataset of publications, the COVID-19 Open Research Dataset (CORD-19). As the pace of research accelerates, biomedical scientists struggle to stay current. To expedite their investigations, scientists leverage hypothesis generation systems, which can automatically inspect published papers to discover novel implicit connections. We present an automated general purpose hypothesis generation systems AGATHA-C and AGATHA-GP for COVID-19 research. The systems are based on graph-mining and the transformer model. The systems are massively validated using retrospective information rediscovery and proactive analysis involving human-in-the-loop expert analysis. Both systems achieve high-quality predictions across domains (in some domains up to 0.97% ROC AUC) in fast computational time and are released to the broad scientific community to accelerate biomedical research. In addition, by performing the domain expert curated study, we show that the systems are able to discover on-going research findings such as the relationship between COVID-19 and oxytocin hormone.
AGATHA: Automatic Graph-mining And Transformer based Hypothesis generation Approach
Feb 13, 2020



Abstract:Medical research is risky and expensive. Drug discovery, as an example, requires that researchers efficiently winnow thousands of potential targets to a small candidate set for more thorough evaluation. However, research groups spend significant time and money to perform the experiments necessary to determine this candidate set long before seeing intermediate results. Hypothesis generation systems address this challenge by mining the wealth of publicly available scientific information to predict plausible research directions. We present AGATHA, a deep-learning hypothesis generation system that can introduce data-driven insights earlier in the discovery process. Through a learned ranking criteria, this system quickly prioritizes plausible term-pairs among entity sets, allowing us to recommend new research directions. We massively validate our system with a temporal holdout wherein we predict connections first introduced after 2015 using data published beforehand. We additionally explore biomedical sub-domains, and demonstrate AGATHA's predictive capacity across the twenty most popular relationship types. This system achieves best-in-class performance on an established benchmark, and demonstrates high recommendation scores across subdomains. Reproducibility: All code, experimental data, and pre-trained models are available online: sybrandt.com/2020/agatha
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge