Huolin L. Xin
Human Perception-Inspired Grain Segmentation Refinement Using Conditional Random Fields
Dec 15, 2023



Abstract:Accurate segmentation of interconnected line networks, such as grain boundaries in polycrystalline material microstructures, poses a significant challenge due to the fragmented masks produced by conventional computer vision algorithms, including convolutional neural networks. These algorithms struggle with thin masks, often necessitating intricate post-processing for effective contour closure and continuity. Addressing this issue, this paper introduces a fast, high-fidelity post-processing technique, leveraging domain knowledge about grain boundary connectivity and employing conditional random fields and perceptual grouping rules. This approach significantly enhances segmentation mask accuracy, achieving a 79% segment identification accuracy in validation with a U-Net model on electron microscopy images of a polycrystalline oxide. Additionally, a novel grain alignment metric is introduced, showing a 51% improvement in grain alignment, providing a more detailed assessment of segmentation performance for complex microstructures. This method not only enables rapid and accurate segmentation but also facilitates an unprecedented level of data analysis, significantly improving the statistical representation of grain boundary networks, making it suitable for a range of disciplines where precise segmentation of interconnected line networks is essential.
Periodic Artifact Reduction in Fourier transforms of Full Field Atomic Resolution Images
Oct 14, 2022Abstract:The discrete Fourier transform is among the most routine tools used in high-resolution scanning / transmission electron microscopy (S/TEM). However, when calculating a Fourier transform, periodic boundary conditions are imposed and sharp discontinuities between the edges of an image cause a cross patterned artifact along the reciprocal space axes. This artifact can interfere with the analysis of reciprocal lattice peaks of an atomic resolution image. Here we demonstrate that the recently developed Periodic Plus Smooth Decomposition technique provides a simple, efficient method for reliable removal of artifacts caused by edge discontinuities. In this method, edge artifacts are reduced by subtracting a smooth background that solves Poisson's equation with boundary conditions set by the image's edges. Unlike the traditional windowed Fourier transforms, Periodic Plus Smooth Decomposition maintains sharp reciprocal lattice peaks from the image's entire field of view.
Electron energy loss spectroscopy database synthesis and automation of core-loss edge recognition by deep-learning neural networks
Sep 26, 2022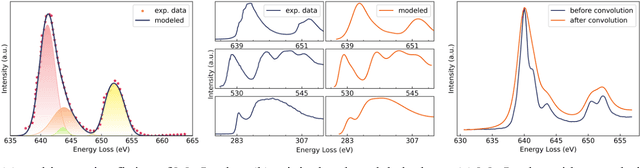

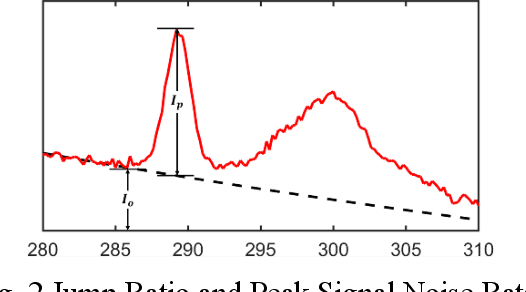
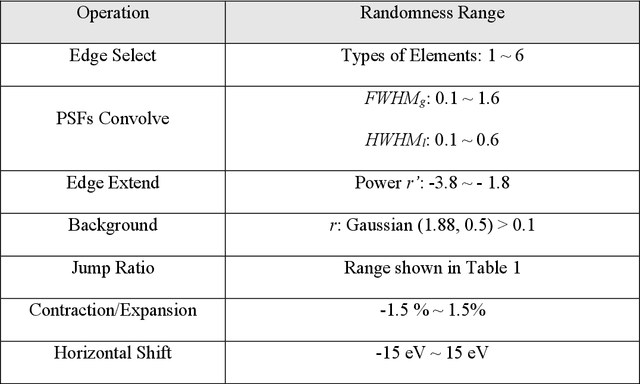
Abstract:The ionization edges encoded in the electron energy loss spectroscopy (EELS) spectra enable advanced material analysis including composition analyses and elemental quantifications. The development of the parallel EELS instrument and fast, sensitive detectors have greatly improved the acquisition speed of EELS spectra. However, the traditional way of core-loss edge recognition is experience based and human labor dependent, which limits the processing speed. So far, the low signal-noise ratio and the low jump ratio of the core-loss edges on the raw EELS spectra have been challenging for the automation of edge recognition. In this work, a convolutional-bidirectional long short-term memory neural network (CNN-BiLSTM) is proposed to automate the detection and elemental identification of core-loss edges from raw spectra. An EELS spectral database is synthesized by using our forward model to assist in the training and validation of the neural network. To make the synthesized spectra resemble the real spectra, we collected a large library of experimentally acquired EELS core edges. In synthesize the training library, the edges are modeled by fitting the multi-gaussian model to the real edges from experiments, and the noise and instrumental imperfectness are simulated and added. The well-trained CNN-BiLSTM network is tested against both the simulated spectra and real spectra collected from experiments. The high accuracy of the network, 94.9 %, proves that, without complicated preprocessing of the raw spectra, the proposed CNN-BiLSTM network achieves the automation of core-loss edge recognition for EELS spectra with high accuracy.
TEMImageNet and AtomSegNet Deep Learning Training Library and Models for High-Precision Atom Segmentation, Localization, Denoising, and Super-resolution Processing of Atom-Resolution Scanning TEM Images
Dec 16, 2020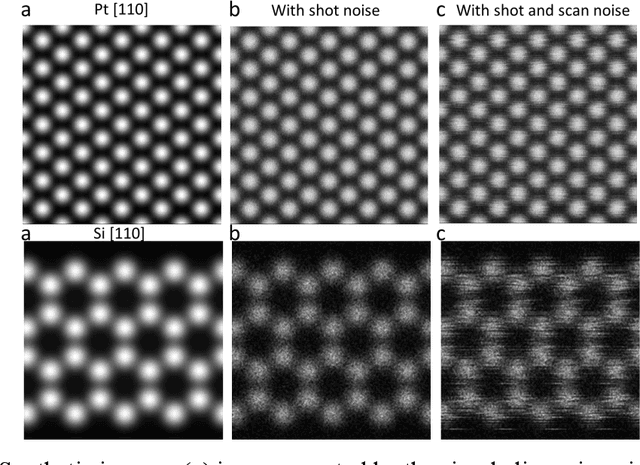

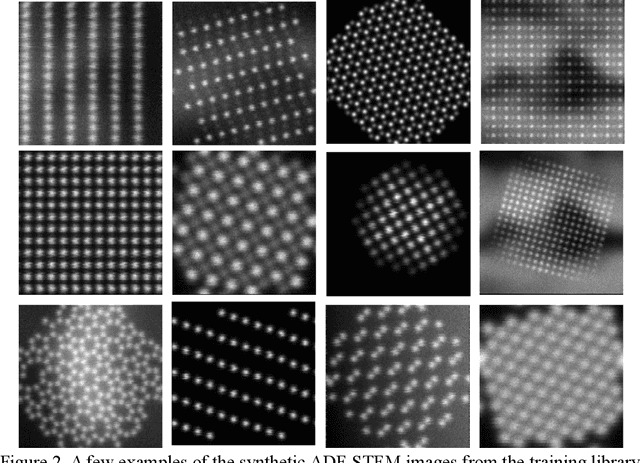
Abstract:Atom segmentation and localization, noise reduction and super-resolution processing of atomic-resolution scanning transmission electron microscopy (STEM) images with high precision and robustness is a challenging task. Although several conventional algorithms, such has thresholding, edge detection and clustering, can achieve reasonable performance in some predefined sceneries, they tend to fail when interferences from the background are strong and unpredictable. Particularly, for atomic-resolution STEM images, so far there is no well-established algorithm that is robust enough to segment or detect all atomic columns when there is large thickness variation in a recorded image. Herein, we report the development of a training library and a deep learning method that can perform robust and precise atom segmentation, localization, denoising, and super-resolution processing of experimental images. Despite using simulated images as training datasets, the deep-learning model can self-adapt to experimental STEM images and shows outstanding performance in atom detection and localization in challenging contrast conditions and the precision is consistently better than the state-of-the-art two-dimensional Gaussian fit method. Taking a step further, we have deployed our deep-learning models to a desktop app with a graphical user interface and the app is free and open-source. We have also built a TEM ImageNet project website for easy browsing and downloading of the training data.
Data Processing For Atomic Resolution EELS
Dec 13, 2011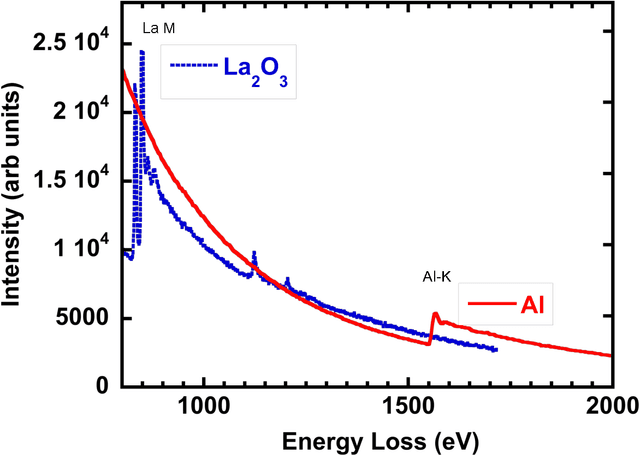
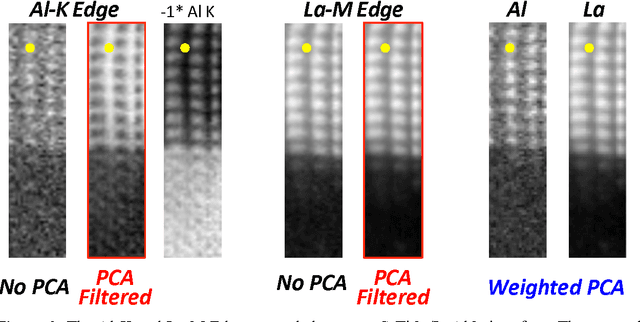
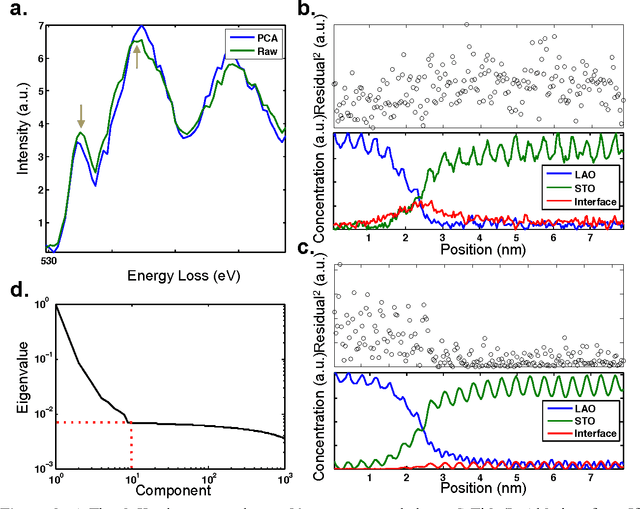
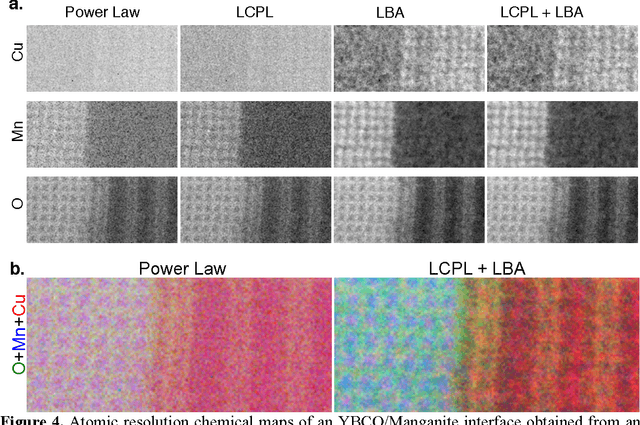
Abstract:The high beam current and sub-angstrom resolution of aberration-corrected scanning transmission electron microscopes has enabled electron energy loss spectroscopic (EELS) mapping with atomic resolution. These spectral maps are often dose-limited and spatially oversampled, leading to low counts/channel and are thus highly sensitive to errors in background estimation. However, by taking advantage of redundancy in the dataset map one can improve background estimation and increase chemical sensitivity. We consider two such approaches- linear combination of power laws and local background averaging-that reduce background error and improve signal extraction. Principal components analysis (PCA) can also be used to analyze spectrum images, but the poor peak-to-background ratio in EELS can lead to serious artifacts if raw EELS data is PCA filtered. We identify common artifacts and discuss alternative approaches. These algorithms are implemented within the Cornell Spectrum Imager, an open source software package for spectroscopic analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge