Huimin Zhu
ActivityDiff: A diffusion model with Positive and Negative Activity Guidance for De Novo Drug Design
Aug 08, 2025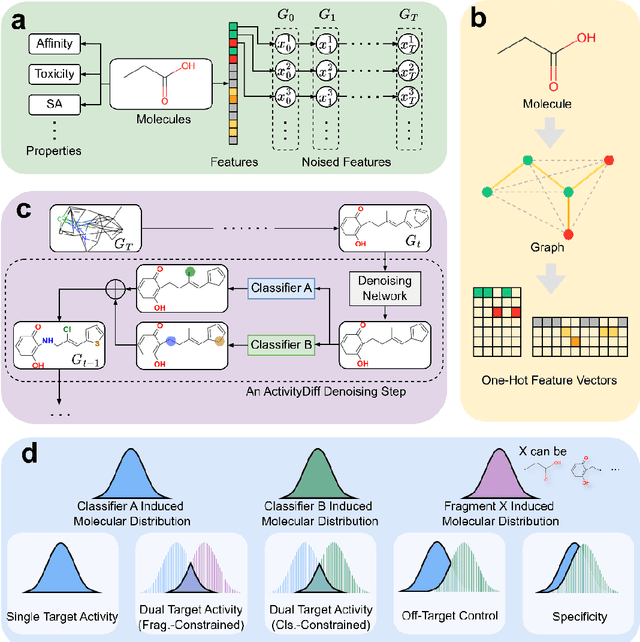
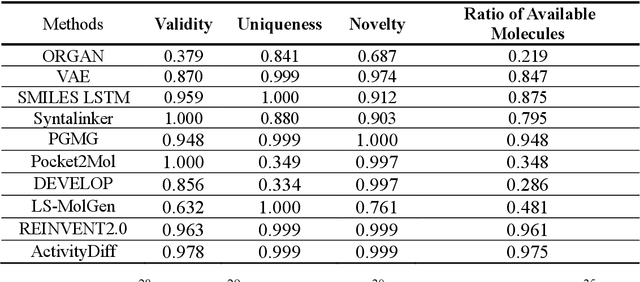
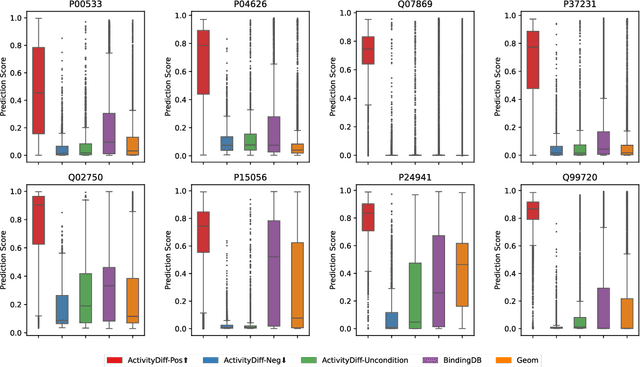
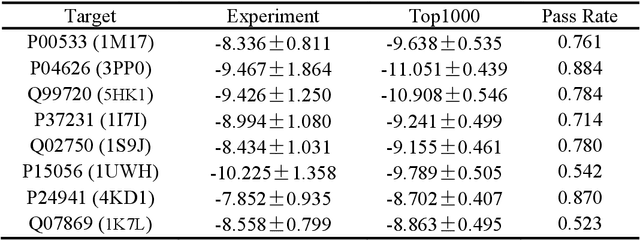
Abstract:Achieving precise control over a molecule's biological activity-encompassing targeted activation/inhibition, cooperative multi-target modulation, and off-target toxicity mitigation-remains a critical challenge in de novo drug design. However, existing generative methods primarily focus on producing molecules with a single desired activity, lacking integrated mechanisms for the simultaneous management of multiple intended and unintended molecular interactions. Here, we propose ActivityDiff, a generative approach based on the classifier-guidance technique of diffusion models. It leverages separately trained drug-target classifiers for both positive and negative guidance, enabling the model to enhance desired activities while minimizing harmful off-target effects. Experimental results show that ActivityDiff effectively handles essential drug design tasks, including single-/dual-target generation, fragment-constrained dual-target design, selective generation to enhance target specificity, and reduction of off-target effects. These results demonstrate the effectiveness of classifier-guided diffusion in balancing efficacy and safety in molecular design. Overall, our work introduces a novel paradigm for achieving integrated control over molecular activity, and provides ActivityDiff as a versatile and extensible framework.
Symbol Detection for Coarsely Quantized OTFS
Sep 21, 2023



Abstract:This paper explicitly models a coarse and noisy quantization in a communication system empowered by orthogonal time frequency space (OTFS) for cost and power efficiency. We first point out, with coarse quantization, the effective channel is imbalanced and thus no longer able to circularly shift the transmitted symbols along the delay-Doppler domain. Meanwhile, the effective channel is non-isotropic, which imposes a significant loss to symbol detection algorithms like the original approximate message passing (AMP). Although the algorithm of generalized expectation consistent for signal recovery (GEC-SR) can mitigate this loss, the complexity in computation is prohibitively high, mainly due to an dramatic increase in the matrix size of OTFS. In this context, we propose a low-complexity algorithm that incorporates into the GEC-SR a quick inversion of quasi-banded matrices, reducing the complexity from a cubic order to a linear order while keeping the performance at the same level.
PGMG: A Pharmacophore-Guided Deep Learning Approach for Bioactive Molecular Generation
Jul 02, 2022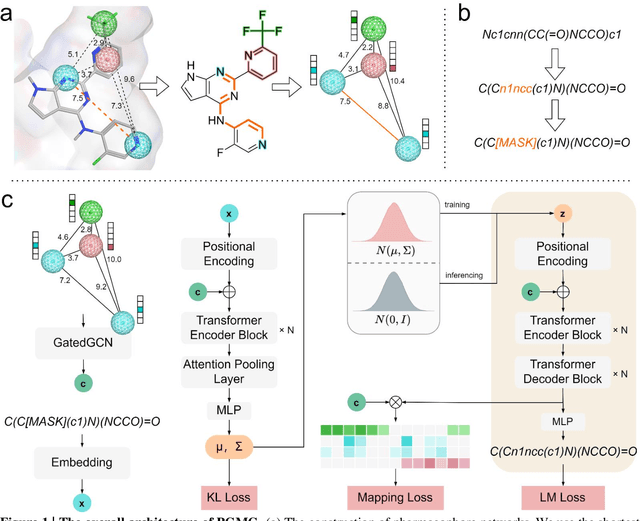
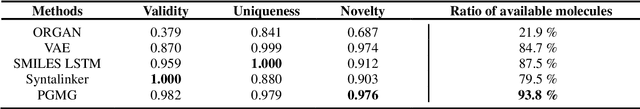
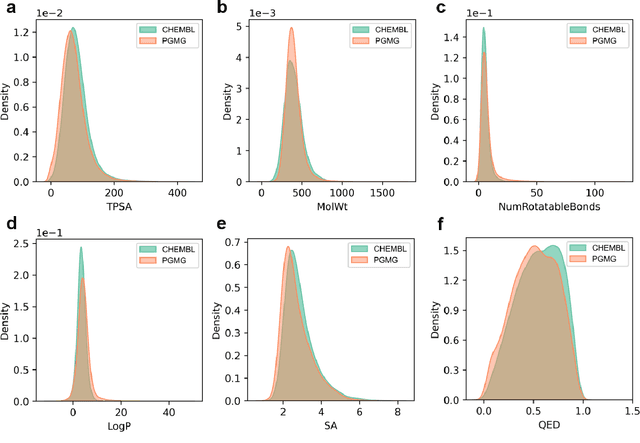
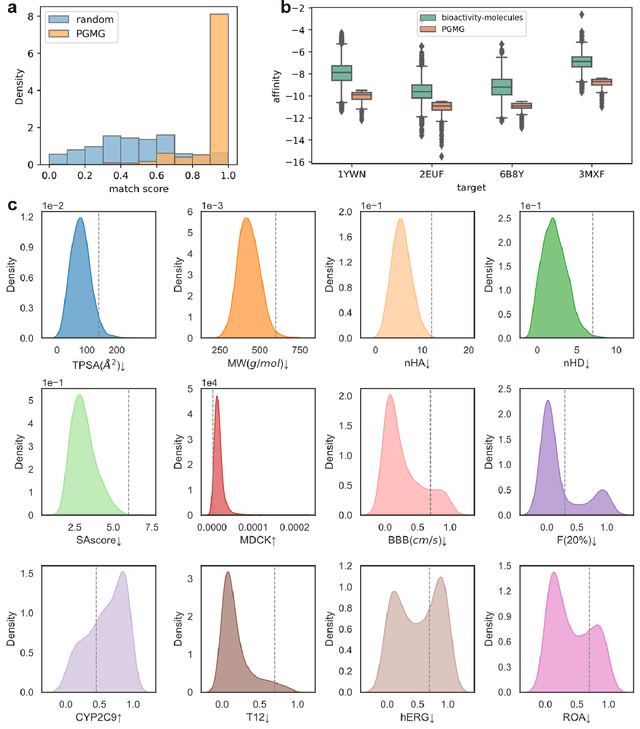
Abstract:The rational design of novel molecules with desired bioactivity is a critical but challenging task in drug discovery, especially when treating a novel target family or understudied targets. Here, we propose PGMG, a pharmacophore-guided deep learning approach for bioactivate molecule generation. Through the guidance of pharmacophore, PGMG provides a flexible strategy to generate bioactive molecules with structural diversity in various scenarios using a trained variational autoencoder. We show that PGMG can generate molecules matching given pharmacophore models while maintaining a high level of validity, uniqueness, and novelty. In the case studies, we demonstrate the application of PGMG to generate bioactive molecules in ligand-based and structure-based drug de novo design, as well as in lead optimization scenarios. Overall, the flexibility and effectiveness of PGMG make it a useful tool for accelerating the drug discovery process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge