Huailiang Peng
RDGCN: Reinforced Dependency Graph Convolutional Network for Aspect-based Sentiment Analysis
Nov 08, 2023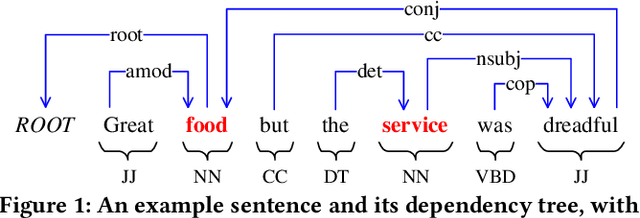
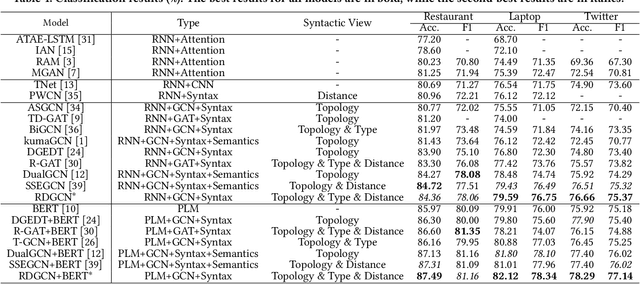
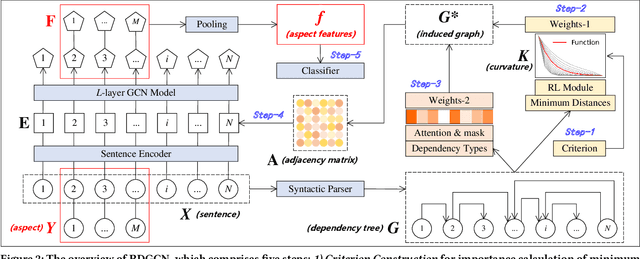
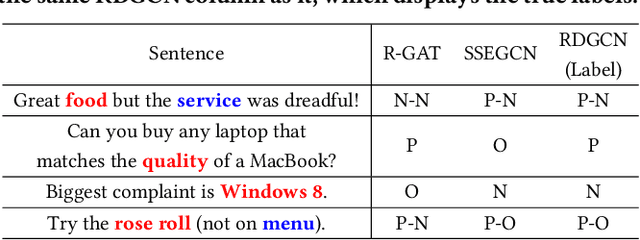
Abstract:Aspect-based sentiment analysis (ABSA) is dedicated to forecasting the sentiment polarity of aspect terms within sentences. Employing graph neural networks to capture structural patterns from syntactic dependency parsing has been confirmed as an effective approach for boosting ABSA. In most works, the topology of dependency trees or dependency-based attention coefficients is often loosely regarded as edges between aspects and opinions, which can result in insufficient and ambiguous syntactic utilization. To address these problems, we propose a new reinforced dependency graph convolutional network (RDGCN) that improves the importance calculation of dependencies in both distance and type views. Initially, we propose an importance calculation criterion for the minimum distances over dependency trees. Under the criterion, we design a distance-importance function that leverages reinforcement learning for weight distribution search and dissimilarity control. Since dependency types often do not have explicit syntax like tree distances, we use global attention and mask mechanisms to design type-importance functions. Finally, we merge these weights and implement feature aggregation and classification. Comprehensive experiments on three popular datasets demonstrate the effectiveness of the criterion and importance functions. RDGCN outperforms state-of-the-art GNN-based baselines in all validations.
Multi-omics Sampling-based Graph Transformer for Synthetic Lethality Prediction
Oct 17, 2023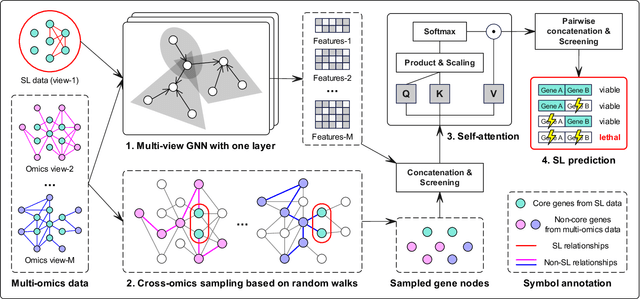
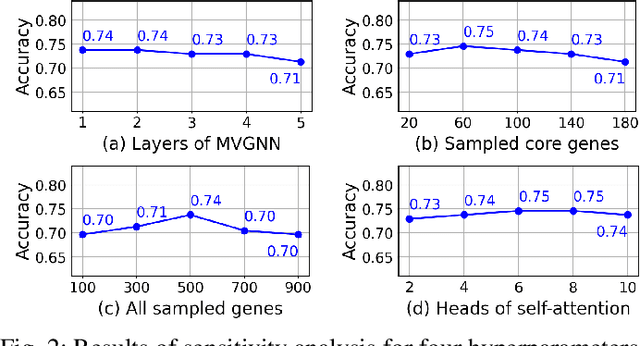
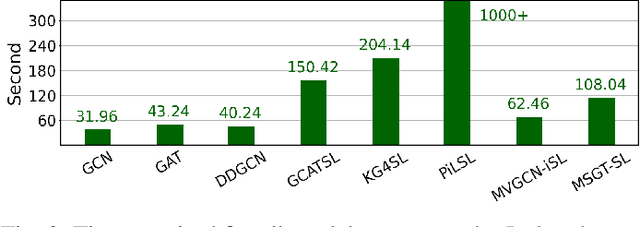
Abstract:Synthetic lethality (SL) prediction is used to identify if the co-mutation of two genes results in cell death. The prevalent strategy is to abstract SL prediction as an edge classification task on gene nodes within SL data and achieve it through graph neural networks (GNNs). However, GNNs suffer from limitations in their message passing mechanisms, including over-smoothing and over-squashing issues. Moreover, harnessing the information of non-SL gene relationships within large-scale multi-omics data to facilitate SL prediction poses a non-trivial challenge. To tackle these issues, we propose a new multi-omics sampling-based graph transformer for SL prediction (MSGT-SL). Concretely, we introduce a shallow multi-view GNN to acquire local structural patterns from both SL and multi-omics data. Further, we input gene features that encode multi-view information into the standard self-attention to capture long-range dependencies. Notably, starting with batch genes from SL data, we adopt parallel random walk sampling across multiple omics gene graphs encompassing them. Such sampling effectively and modestly incorporates genes from omics in a structure-aware manner before using self-attention. We showcase the effectiveness of MSGT-SL on real-world SL tasks, demonstrating the empirical benefits gained from the graph transformer and multi-omics data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge