Hongchao Qin
OpenMAG: A Comprehensive Benchmark for Multimodal-Attributed Graph
Feb 05, 2026Abstract:Multimodal-Attributed Graph (MAG) learning has achieved remarkable success in modeling complex real-world systems by integrating graph topology with rich attributes from multiple modalities. With the rapid proliferation of novel MAG models capable of handling intricate cross-modal semantics and structural dependencies, establishing a rigorous and unified evaluation standard has become imperative. Although existing benchmarks have facilitated initial progress, they exhibit critical limitations in domain coverage, encoder flexibility, model diversity, and task scope, presenting significant challenges to fair evaluation. To bridge this gap, we present OpenMAG, a comprehensive benchmark that integrates 19 datasets across 6 domains and incorporates 16 encoders to support both static and trainable feature encoding. OpenMAG further implements a standardized library of 24 state-of-the-art models and supports 8 downstream tasks, enabling fair comparisons within a unified framework. Through systematic assessment of necessity, data quality, effectiveness, robustness, and efficiency, we derive 14 fundamental insights into MAG learning to guide future advancements. Our code is available at https://github.com/YUKI-N810/OpenMAG.
Toward Effective Multimodal Graph Foundation Model: A Divide-and-Conquer Based Approach
Feb 04, 2026Abstract:Graph Foundation Models (GFMs) have achieved remarkable success in generalizing across diverse domains. However, they mainly focus on Text-Attributed Graphs (TAGs), leaving Multimodal-Attributed Graphs (MAGs) largely untapped. Developing Multimodal Graph Foundation Models (MGFMs) allows for leveraging the rich multimodal information in MAGs, and extends applicability to broader types of downstream tasks. While recent MGFMs integrate diverse modality information, our empirical investigation reveals two fundamental limitations of existing MGFMs: (1)they fail to explicitly model modality interaction, essential for capturing intricate cross-modal semantics beyond simple aggregation, and (2)they exhibit sub-optimal modality alignment, which is critical for bridging the significant semantic disparity between distinct modal spaces. To address these challenges, we propose PLANET (graPh topoLogy-aware modAlity iNteraction and alignmEnT), a novel framework employing a Divide-and-Conquer strategy to decouple modality interaction and alignment across distinct granularities. At the embedding granularity, (1)Embedding-wise Domain Gating (EDG) performs local semantic enrichment by adaptively infusing topology-aware cross-modal context, achieving modality interaction. At the node granularity, (2)Node-wise Discretization Retrieval (NDR) ensures global modality alignment by constructing a Discretized Semantic Representation Space (DSRS) to bridge modality gaps. Extensive experiments demonstrate that PLANET significantly outperforms state-of-the-art baselines across diverse graph-centric and multimodal generative tasks.
DOGMA: Weaving Structural Information into Data-centric Single-cell Transcriptomics Analysis
Feb 02, 2026Abstract:Recently, data-centric AI methodology has been a dominant paradigm in single-cell transcriptomics analysis, which treats data representation rather than model complexity as the fundamental bottleneck. In the review of current studies, earlier sequence methods treat cells as independent entities and adapt prevalent ML models to analyze their directly inherited sequence data. Despite their simplicity and intuition, these methods overlook the latent intercellular relationships driven by the functional mechanisms of biological systems and the inherent quality issues of the raw sequence data. Therefore, a series of structured methods has emerged. Although they employ various heuristic rules to capture intricate intercellular relationships and enhance the raw sequencing data, these methods often neglect biological prior knowledge. This omission incurs substantial overhead and yields suboptimal graph representations, thereby hindering the utility of ML models. To address them, we propose DOGMA, a holistic data-centric framework designed for the structural reshaping and semantic enhancement of raw data through multi-level biological prior knowledge. Transcending reliance on stochastic heuristics, DOGMA redefines graph construction by integrating Statistical Anchors with Cell Ontology and Phylogenetic Trees to enable deterministic structure discovery and robust cross-species alignment. Furthermore, Gene Ontology is utilized to bridge the feature-level semantic gap by incorporating functional priors. In complex multi-species and multi-organ benchmarks, DOGMA achieves SOTA performance, exhibiting superior zero-shot robustness and sample efficiency while operating with significantly lower computational cost.
OpenDDI: A Comprehensive Benchmark for DDI Prediction
Jan 31, 2026Abstract:Drug-Drug Interactions (DDIs) significantly influence therapeutic efficacy and patient safety. As experimental discovery is resource-intensive and time-consuming, efficient computational methodologies have become essential. The predominant paradigm formulates DDI prediction as a drug graph-based link prediction task. However, further progress is hindered by two fundamental challenges: (1) lack of high-quality data: most studies rely on small-scale DDI datasets and single-modal drug representations; (2) lack of standardized evaluation: inconsistent scenarios, varied metrics, and diverse baselines. To address the above issues, we propose OpenDDI, a comprehensive benchmark for DDI prediction. Specifically, (1) from the data perspective, OpenDDI unifies 6 widely used DDI datasets and 2 existing forms of drug representation, while additionally contributing 3 new large-scale LLM-augmented datasets and a new multimodal drug representation covering 5 modalities. (2) From the evaluation perspective, OpenDDI unifies 20 SOTA model baselines across 3 downstream tasks, with standardized protocols for data quality, effectiveness, generalization, robustness, and efficiency. Based on OpenDDI, we conduct a comprehensive evaluation and derive 10 valuable insights for DDI prediction while exposing current limitations to provide critical guidance for this rapidly evolving field. Our code is available at https://github.com/xiaoriwuguang/OpenDDI
Scalable Topology-Preserving Graph Coarsening with Graph Collapse
Jan 30, 2026Abstract:Graph coarsening reduces the size of a graph while preserving certain properties. Most existing methods preserve either spectral or spatial characteristics. Recent research has shown that preserving topological features helps maintain the predictive performance of graph neural networks (GNNs) trained on the coarsened graph but suffers from exponential time complexity. To address these problems, we propose Scalable Topology-Preserving Graph Coarsening (STPGC) by introducing the concepts of graph strong collapse and graph edge collapse extended from algebraic topology. STPGC comprises three new algorithms, GStrongCollapse, GEdgeCollapse, and NeighborhoodConing based on these two concepts, which eliminate dominated nodes and edges while rigorously preserving topological features. We further prove that STPGC preserves the GNN receptive field and develop approximate algorithms to accelerate GNN training. Experiments on node classification with GNNs demonstrate the efficiency and effectiveness of STPGC.
LION: A Clifford Neural Paradigm for Multimodal-Attributed Graph Learning
Jan 29, 2026Abstract:Recently, the rapid advancement of multimodal domains has driven a data-centric paradigm shift in graph ML, transitioning from text-attributed to multimodal-attributed graphs. This advancement significantly enhances data representation and expands the scope of graph downstream tasks, such as modality-oriented tasks, thereby improving the practical utility of graph ML. Despite its promise, limitations exist in the current neural paradigms: (1) Neglect Context in Modality Alignment: Most existing methods adopt topology-constrained or modality-specific operators as tokenizers. These aligners inevitably neglect graph context and inhibit modality interaction, resulting in suboptimal alignment. (2) Lack of Adaptation in Modality Fusion: Most existing methods are simple adaptations for 2-modality graphs and fail to adequately exploit aligned tokens equipped with topology priors during fusion, leading to poor generalizability and performance degradation. To address the above issues, we propose LION (c\underline{LI}ff\underline{O}rd \underline{N}eural paradigm) based on the Clifford algebra and decoupled graph neural paradigm (i.e., propagation-then-aggregation) to implement alignment-then-fusion in multimodal-attributed graphs. Specifically, we first construct a modality-aware geometric manifold grounded in Clifford algebra. This geometric-induced high-order graph propagation efficiently achieves modality interaction, facilitating modality alignment. Then, based on the geometric grade properties of aligned tokens, we propose adaptive holographic aggregation. This module integrates the energy and scale of geometric grades with learnable parameters to improve modality fusion. Extensive experiments on 9 datasets demonstrate that LION significantly outperforms SOTA baselines across 3 graph and 3 modality downstream tasks.
DANCE: Dynamic, Available, Neighbor-gated Condensation for Federated Text-Attributed Graphs
Jan 23, 2026Abstract:Federated graph learning (FGL) enables collaborative training on graph data across multiple clients. With the rise of large language models (LLMs), textual attributes in FGL graphs are gaining attention. Text-attributed graph federated learning (TAG-FGL) improves FGL by explicitly leveraging LLMs to process and integrate these textual features. However, current TAG-FGL methods face three main challenges: \textbf{(1) Overhead.} LLMs for processing long texts incur high token and computation costs. To make TAG-FGL practical, we introduce graph condensation (GC) to reduce computation load, but this choice also brings new issues. \textbf{(2) Suboptimal.} To reduce LLM overhead, we introduce GC into TAG-FGL by compressing multi-hop texts/neighborhoods into a condensed core with fixed LLM surrogates. However, this one-shot condensation is often not client-adaptive, leading to suboptimal performance. \textbf{(3) Interpretability.} LLM-based condensation further introduces a black-box bottleneck: summaries lack faithful attribution and clear grounding to specific source spans, making local inspection and auditing difficult. To address the above issues, we propose \textbf{DANCE}, a new TAG-FGL paradigm with GC. To improve \textbf{suboptimal} performance, DANCE performs round-wise, model-in-the-loop condensation refresh using the latest global model. To enhance \textbf{interpretability}, DANCE preserves provenance by storing locally inspectable evidence packs that trace predictions to selected neighbors and source text spans. Across 8 TAG datasets, DANCE improves accuracy by \textbf{2.33\%} at an \textbf{8\%} condensation ratio, with \textbf{33.42\%} fewer tokens than baselines.
Toward Data-centric Directed Graph Learning: An Entropy-driven Approach
May 02, 2025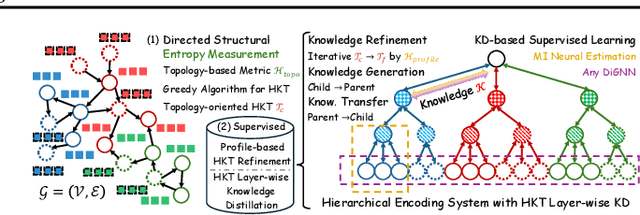
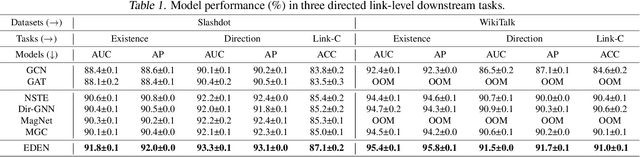
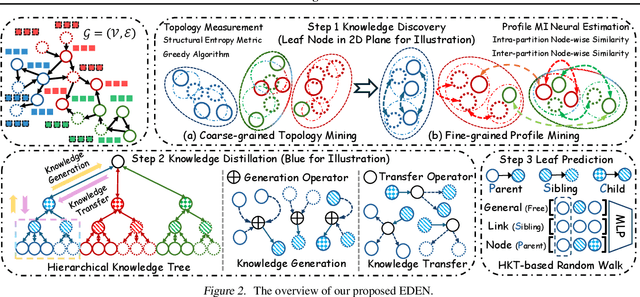
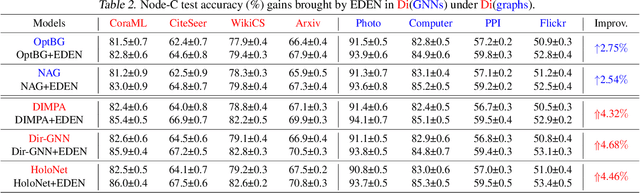
Abstract:The directed graph (digraph), as a generalization of undirected graphs, exhibits superior representation capability in modeling complex topology systems and has garnered considerable attention in recent years. Despite the notable efforts made by existing DiGraph Neural Networks (DiGNNs) to leverage directed edges, they still fail to comprehensively delve into the abundant data knowledge concealed in the digraphs. This data-level limitation results in model-level sub-optimal predictive performance and underscores the necessity of further exploring the potential correlations between the directed edges (topology) and node profiles (feature and labels) from a data-centric perspective, thereby empowering model-centric neural networks with stronger encoding capabilities. In this paper, we propose \textbf{E}ntropy-driven \textbf{D}igraph knowl\textbf{E}dge distillatio\textbf{N} (EDEN), which can serve as a data-centric digraph learning paradigm or a model-agnostic hot-and-plug data-centric Knowledge Distillation (KD) module. The core idea is to achieve data-centric ML, guided by our proposed hierarchical encoding theory for structured data. Specifically, EDEN first utilizes directed structural measurements from a topology perspective to construct a coarse-grained Hierarchical Knowledge Tree (HKT). Subsequently, EDEN quantifies the mutual information of node profiles to refine knowledge flow in the HKT, enabling data-centric KD supervision within model training. As a general framework, EDEN can also naturally extend to undirected scenarios and demonstrate satisfactory performance. In our experiments, EDEN has been widely evaluated on 14 (di)graph datasets (homophily and heterophily) and across 4 downstream tasks. The results demonstrate that EDEN attains SOTA performance and exhibits strong improvement for prevalent (Di)GNNs.
Rethinking Graph Structure Learning in the Era of LLMs
Mar 27, 2025



Abstract:Recently, the emergence of large language models (LLMs) has prompted researchers to explore the integration of language descriptions into graphs, aiming to enhance model encoding capabilities from a data-centric perspective. This graph representation is called text-attributed graphs (TAGs). A review of prior advancements highlights that graph structure learning (GSL) is a pivotal technique for improving data utility, making it highly relevant to efficient TAG learning. However, most GSL methods are tailored for traditional graphs without textual information, underscoring the necessity of developing a new GSL paradigm. Despite clear motivations, it remains challenging: (1) How can we define a reasonable optimization objective for GSL in the era of LLMs, considering the massive parameters in LLM? (2) How can we design an efficient model architecture that enables seamless integration of LLM for this optimization objective? For Question 1, we reformulate existing GSL optimization objectives as a tree optimization framework, shifting the focus from obtaining a well-trained edge predictor to a language-aware tree sampler. For Question 2, we propose decoupled and training-free model design principles for LLM integration, shifting the focus from computation-intensive fine-tuning to more efficient inference. Based on this, we propose Large Language and Tree Assistant (LLaTA), which leverages tree-based LLM in-context learning to enhance the understanding of topology and text, enabling reliable inference and generating improved graph structure. Extensive experiments on 10 TAG datasets demonstrate that LLaTA enjoys flexibility - incorporated with any backbone; scalability - outperforms other LLM-based GSL methods in terms of running efficiency; effectiveness - achieves SOTA performance.
Toward Effective Digraph Representation Learning: A Magnetic Adaptive Propagation based Approach
Jan 21, 2025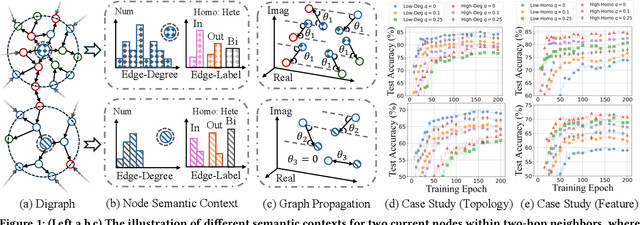
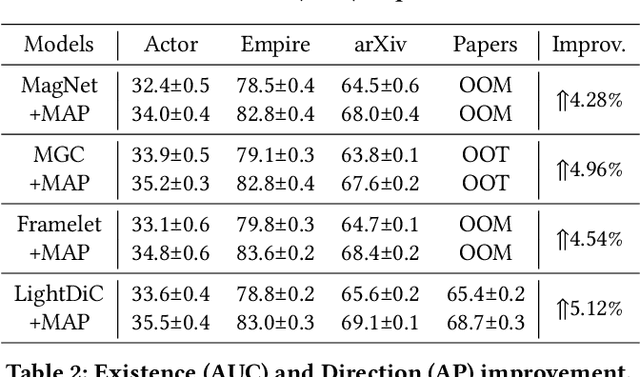
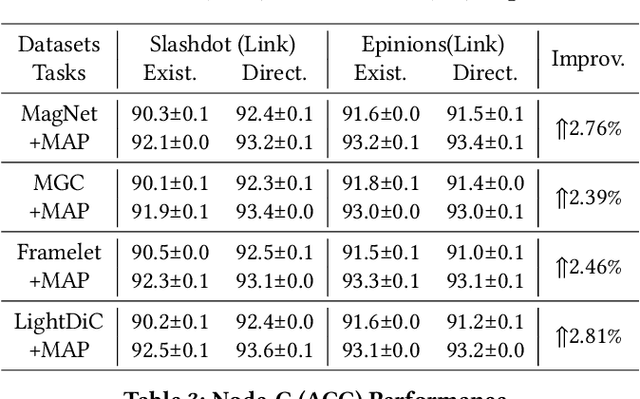
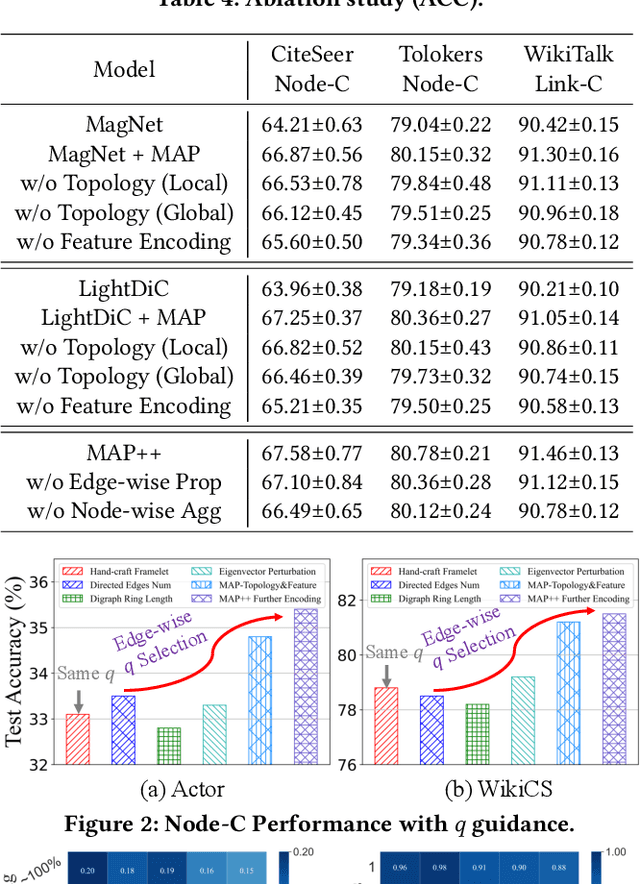
Abstract:The $q$-parameterized magnetic Laplacian serves as the foundation of directed graph (digraph) convolution, enabling this kind of digraph neural network (MagDG) to encode node features and structural insights by complex-domain message passing. As a generalization of undirected methods, MagDG shows superior capability in modeling intricate web-scale topology. Despite the great success achieved by existing MagDGs, limitations still exist: (1) Hand-crafted $q$: The performance of MagDGs depends on selecting an appropriate $q$-parameter to construct suitable graph propagation equations in the complex domain. This parameter tuning, driven by downstream tasks, limits model flexibility and significantly increases manual effort. (2) Coarse Message Passing: Most approaches treat all nodes with the same complex-domain propagation and aggregation rules, neglecting their unique digraph contexts. This oversight results in sub-optimal performance. To address the above issues, we propose two key techniques: (1) MAP is crafted to be a plug-and-play complex-domain propagation optimization strategy in the context of digraph learning, enabling seamless integration into any MagDG to improve predictions while enjoying high running efficiency. (2) MAP++ is a new digraph learning framework, further incorporating a learnable mechanism to achieve adaptively edge-wise propagation and node-wise aggregation in the complex domain for better performance. Extensive experiments on 12 datasets demonstrate that MAP enjoys flexibility for it can be incorporated with any MagDG, and scalability as it can deal with web-scale digraphs. MAP++ achieves SOTA predictive performance on 4 different downstream tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge