Hatim Aboalsamh
A Deep Learning-Based Unified Framework for Red Lesions Detection on Retinal Fundus Images
Sep 17, 2021
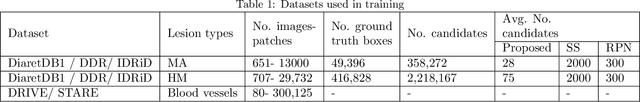

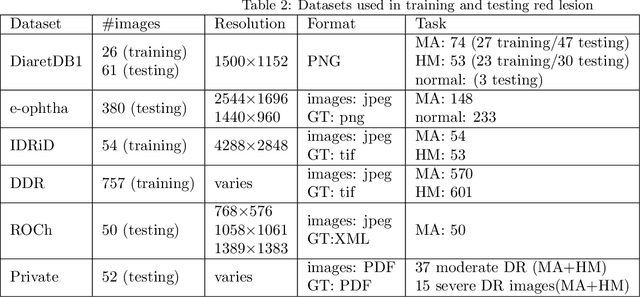
Abstract:Red-lesions, i.e., microaneurysms (MAs) and hemorrhages (HMs), are the early signs of diabetic retinopathy (DR). The automatic detection of MAs and HMs on retinal fundus images is a challenging task. Most of the existing methods detect either only MAs or only HMs because of the difference in their texture, sizes, and morphology. Though some methods detect both MAs and HMs, they suffer from the curse of dimensionality of shape and colors features and fail to detect all shape variations of HMs such as flame-shaped HM. Leveraging the progress in deep learning, we proposed a two-stream red lesions detection system dealing simultaneously with small and large red lesions. For this system, we introduced a new ROIs candidates generation method for large red lesions fundus images; it is based on blood vessel segmentation and morphological operations, and reduces the computational complexity, and enhances the detection accuracy by generating a small number of potential candidates. For detection, we adapted the Faster RCNN framework with two streams. We used pre-trained VGGNet as a bone model and carried out several extensive experiments to tune it for vessels segmentation and candidates generation, and finally learning the appropriate mapping, which yields better detection of the red lesions comparing with the state-of-the-art methods. The experimental results validated the effectiveness of the system in the detection of both MAs and HMs; the method yields higher performance for per lesion detection according to sensitivity under 4 FPIs on DiaretDB1-MA and DiaretDB1-HM datasets, and 1 FPI on e-ophtha and ROCh datasets than the state of the art methods w.r.t. various evaluation metrics. For DR screening, the system outperforms other methods on DiaretDB1-MA, DiaretDB1-HM, and e-ophtha datasets.
An Efficient Intelligent System for the Classification of Electroencephalography (EEG) Brain Signals using Nuclear Features for Human Cognitive Tasks
Apr 30, 2019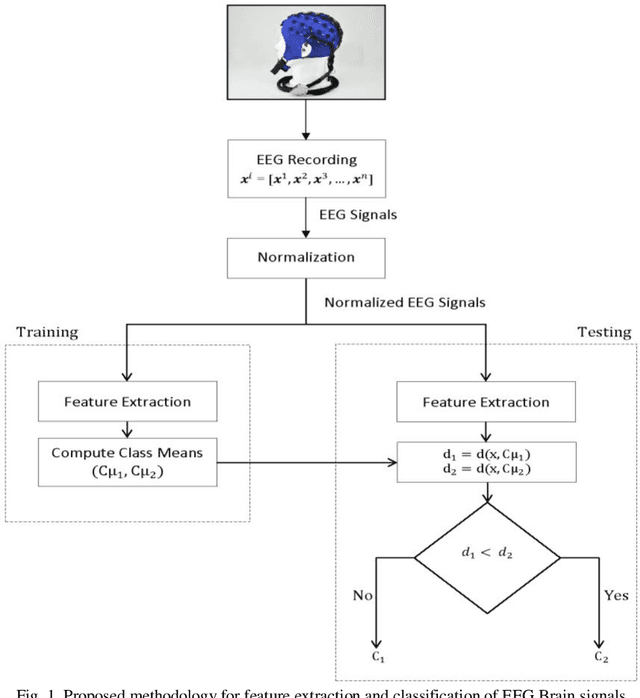
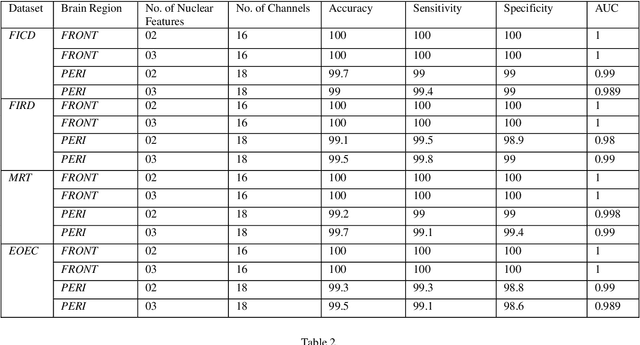
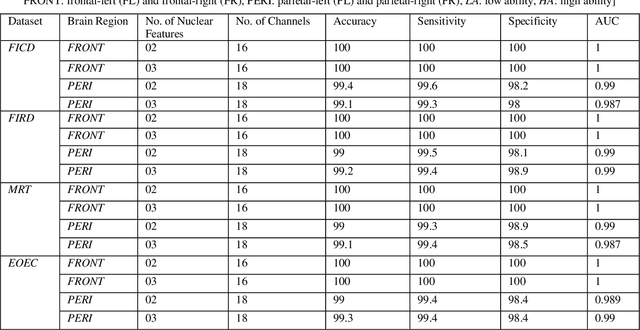
Abstract:Representation and classification of Electroencephalography (EEG) brain signals are critical processes for their analysis in cognitive tasks. Particularly, extraction of discriminative features from raw EEG signals, without any pre-processing, is a challenging task. Motivated by nuclear norm, we observed that there is a significant difference between the variances of EEG signals captured from the same brain region when a subject performs different tasks. This observation lead us to use singular value decomposition for computing dominant variances of EEG signals captured from a certain brain region while performing a certain task and use them as features (nuclear features). A simple and efficient class means based minimum distance classifier (CMMDC) is enough to predict brain states. This approach results in the feature space of significantly small dimension and gives equally good classification results on clean as well as raw data. We validated the effectiveness and robustness of the technique using four datasets of different tasks: fluid intelligence clean data (FICD), fluid intelligence raw data (FIRD), memory recall task (MRT), and eyes open / eyes closed task (EOEC). For each task, we analyzed EEG signals over six (06) different brain regions with 8, 16, 20, 18, 18 and 100 electrodes. The nuclear features from frontal brain region gave the 100% prediction accuracy. The discriminant analysis of the nuclear features has been conducted using intra-class and inter-class variations. Comparisons with the state-of-the-art techniques showed the superiority of the proposed system.
Eigen Values Features for the Classification of Brain Signals corresponding to 2D and 3D Educational Contents
Apr 30, 2019



Abstract:In this paper, we have proposed a brain signal classification method, which uses eigenvalues of the covariance matrix as features to classify images (topomaps) created from the brain signals. The signals are recorded during the answering of 2D and 3D questions. The system is used to classify the correct and incorrect answers for both 2D and 3D questions. Using the classification technique, the impacts of 2D and 3D multimedia educational contents on learning, memory retention and recall will be compared. The subjects learn similar 2D and 3D educational contents. Afterwards, subjects are asked 20 multiple-choice questions (MCQs) associated with the contents after thirty minutes (Short-Term Memory) and two months (Long-Term Memory). Eigenvalues features extracted from topomaps images are given to K-Nearest Neighbor (KNN) and Support Vector Machine (SVM) classifiers, in order to identify the states of the brain related to incorrect and correct answers. Excellent accuracies obtained by both classifiers and by applying statistical analysis on the results, no significant difference is indicated between 2D and 3D multimedia educational contents on learning, memory retention and recall in both STM and LTM.
ConvSRC: SmartPhone based Periocular Recognition using Deep Convolutional Neural Network and Sparsity Augmented Collaborative Representation
Jan 16, 2018

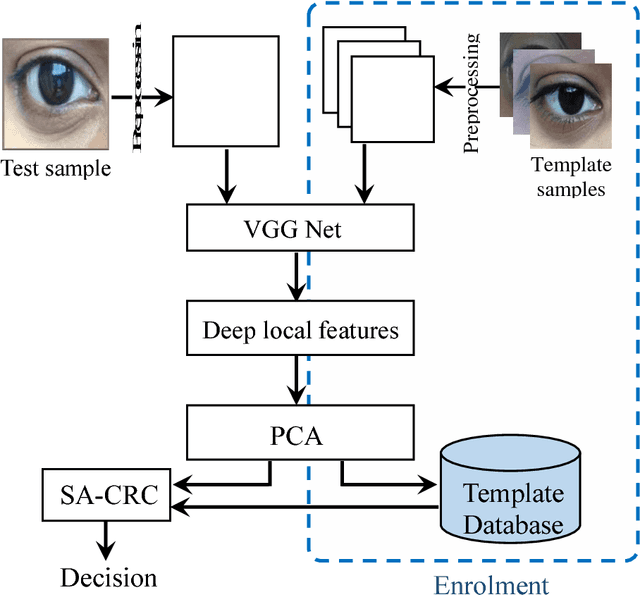
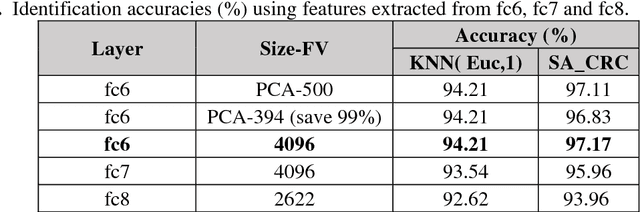
Abstract:Smartphone based periocular recognition has gained significant attention from biometric research community because of the limitations of biometric modalities like face, iris etc. Most of the existing methods for periocular recognition employ hand-crafted features. Recently, learning based image representation techniques like deep Convolutional Neural Network (CNN) have shown outstanding performance in many visual recognition tasks. CNN needs a huge volume of data for its learning, but for periocular recognition only limited amount of data is available. The solution is to use CNN pre-trained on the dataset from the related domain, in this case the challenge is to extract efficiently the discriminative features. Using a pertained CNN model (VGG-Net), we propose a simple, efficient and compact image representation technique that takes into account the wealth of information and sparsity existing in the activations of the convolutional layers and employs principle component analysis. For recognition, we use an efficient and robust Sparse Augmented Collaborative Representation based Classification (SA-CRC) technique. For thorough evaluation of ConvSRC (the proposed system), experiments were carried out on the VISOB challenging database which was presented for periocular recognition competition in ICIP2016. The obtained results show the superiority of ConvSRC over the state-of-the-art methods; it obtains a GMR of more than 99% at FMR = 10-3 and outperforms the first winner of ICIP2016 challenge by 10%.
An Automated System for Epilepsy Detection using EEG Brain Signals based on Deep Learning Approach
Jan 16, 2018



Abstract:Epilepsy is a neurological disorder and for its detection, encephalography (EEG) is a commonly used clinical approach. Manual inspection of EEG brain signals is a time-consuming and laborious process, which puts heavy burden on neurologists and affects their performance. Several automatic techniques have been proposed using traditional approaches to assist neurologists in detecting binary epilepsy scenarios e.g. seizure vs. non-seizure or normal vs. ictal. These methods do not perform well when classifying ternary case e.g. ictal vs. normal vs. inter-ictal; the maximum accuracy for this case by the state-of-the-art-methods is 97+-1%. To overcome this problem, we propose a system based on deep learning, which is an ensemble of pyramidal one-dimensional convolutional neural network (P-1D-CNN) models. In a CNN model, the bottleneck is the large number of learnable parameters. P-1D-CNN works on the concept of refinement approach and it results in 60% fewer parameters compared to traditional CNN models. Further to overcome the limitations of small amount of data, we proposed augmentation schemes for learning P-1D-CNN model. In almost all the cases concerning epilepsy detection, the proposed system gives an accuracy of 99.1+-0.9% on the University of Bonn dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge