Hannah Thompson
Deep learning classifier of locally advanced rectal cancer treatment response from endoscopy images
May 06, 2024
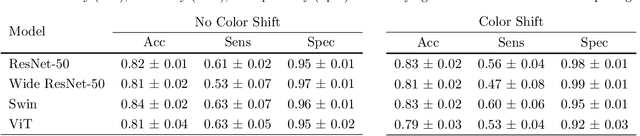
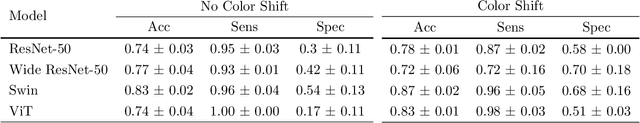

Abstract:We developed a deep learning classifier of rectal cancer response (tumor vs. no-tumor) to total neoadjuvant treatment (TNT) from endoscopic images acquired before, during, and following TNT. We further evaluated the network's ability in a near out-of-distribution (OOD) problem to identify local regrowth (LR) from follow-up endoscopy images acquired several months to years after completing TNT. We addressed endoscopic image variability by using optimal mass transport-based image harmonization. We evaluated multiple training regularization schemes to study the ResNet-50 network's in-distribution and near-OOD generalization ability. Test time augmentation resulted in the most considerable accuracy improvement. Image harmonization resulted in slight accuracy improvement for the near-OOD cases. Our results suggest that off-the-shelf deep learning classifiers can detect rectal cancer from endoscopic images at various stages of therapy for surveillance.
A Framework for Implementing Machine Learning on Omics Data
Nov 26, 2018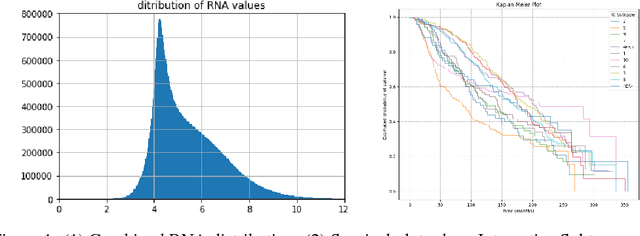
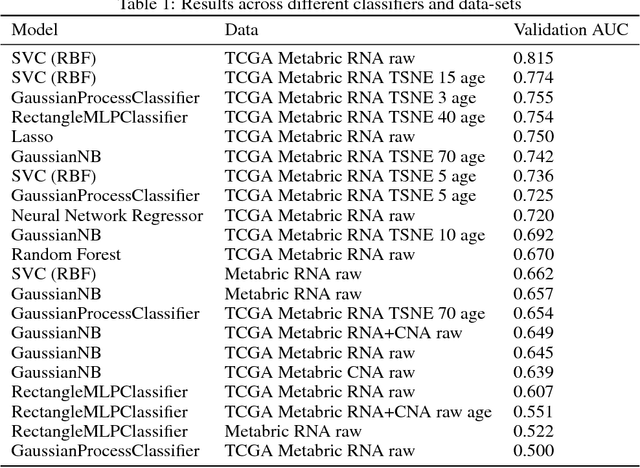
Abstract:The potential benefits of applying machine learning methods to -omics data are becoming increasingly apparent, especially in clinical settings. However, the unique characteristics of these data are not always well suited to machine learning techniques. These data are often generated across different technologies in different labs, and frequently with high dimensionality. In this paper we present a framework for combining -omics data sets, and for handling high dimensional data, making -omics research more accessible to machine learning applications. We demonstrate the success of this framework through integration and analysis of multi-analyte data for a set of 3,533 breast cancers. We then use this data-set to predict breast cancer patient survival for individuals at risk of an impending event, with higher accuracy and lower variance than methods trained on individual data-sets. We hope that our pipelines for data-set generation and transformation will open up -omics data to machine learning researchers. We have made these freely available for noncommercial use at www.ccg.ai.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge