Guozheng Wei
Seq-SetNet: Exploring Sequence Sets for Inferring Structures
Jun 06, 2019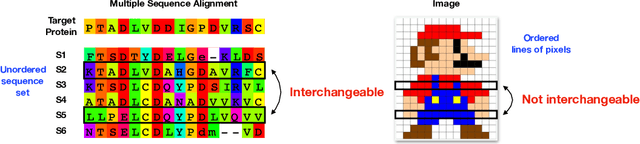
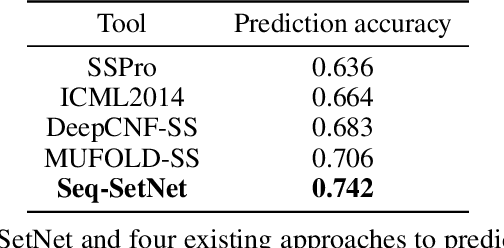
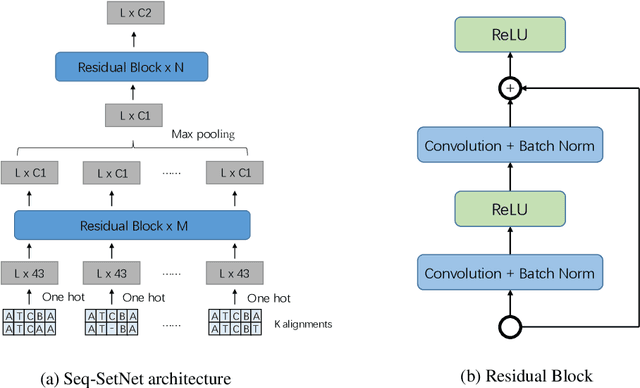
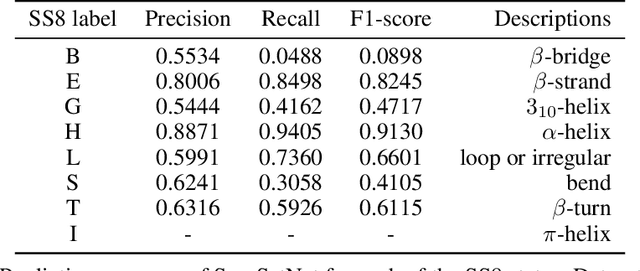
Abstract:Sequence set is a widely-used type of data source in a large variety of fields. A typical example is protein structure prediction, which takes an multiple sequence alignment (MSA) as input and aims to infer structural information from it. Almost all of the existing approaches exploit MSAs in an indirect fashion, i.e., they transform MSAs into position-specific scoring matrices (PSSM) that represent the distribution of amino acid types at each column. PSSM could capture column-wise characteristics of MSA, however, the column-wise characteristics embedded in each individual component sequence were nearly totally neglected. The drawback of PSSM is rooted in the fact that an MSA is essentially an unordered sequence set rather than a matrix. Specifically, the interchange of any two sequences will not affect the whole MSA. In contrast, the pixels in an image essentially form a matrix since any two rows of pixels cannot be interchanged. Therefore, the traditional deep neural networks designed for image processing cannot be directly applied on sequence sets. Here, we proposed a novel deep neural network framework (called Seq-SetNet) for sequence set processing. By employing a {\it symmetric function} module to integrate features calculated from preceding layers, Seq-SetNet are immune to the order of sequences in the input MSA. This advantage enables us to directly and fully exploit MSAs by considering each component protein individually. We evaluated Seq-SetNet by using it to extract structural information from MSA for protein secondary structure prediction. Experimental results on popular benchmark sets suggests that Seq-SetNet outperforms the state-of-the-art approaches by 3.6% in precision. These results clearly suggest the advantages of Seq-SetNet in sequence set processing and it can be readily used in a wide range of fields, say natural language processing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge