Gulshan Saleem
A Robust Deep Networks based Multi-Object MultiCamera Tracking System for City Scale Traffic
May 01, 2025Abstract:Vision sensors are becoming more important in Intelligent Transportation Systems (ITS) for traffic monitoring, management, and optimization as the number of network cameras continues to rise. However, manual object tracking and matching across multiple non-overlapping cameras pose significant challenges in city-scale urban traffic scenarios. These challenges include handling diverse vehicle attributes, occlusions, illumination variations, shadows, and varying video resolutions. To address these issues, we propose an efficient and cost-effective deep learning-based framework for Multi-Object Multi-Camera Tracking (MO-MCT). The proposed framework utilizes Mask R-CNN for object detection and employs Non-Maximum Suppression (NMS) to select target objects from overlapping detections. Transfer learning is employed for re-identification, enabling the association and generation of vehicle tracklets across multiple cameras. Moreover, we leverage appropriate loss functions and distance measures to handle occlusion, illumination, and shadow challenges. The final solution identification module performs feature extraction using ResNet-152 coupled with Deep SORT based vehicle tracking. The proposed framework is evaluated on the 5th AI City Challenge dataset (Track 3), comprising 46 camera feeds. Among these 46 camera streams, 40 are used for model training and validation, while the remaining six are utilized for model testing. The proposed framework achieves competitive performance with an IDF1 score of 0.8289, and precision and recall scores of 0.9026 and 0.8527 respectively, demonstrating its effectiveness in robust and accurate vehicle tracking.
Image Quality Assessment for Foliar Disease Identification (AgroPath)
Sep 26, 2022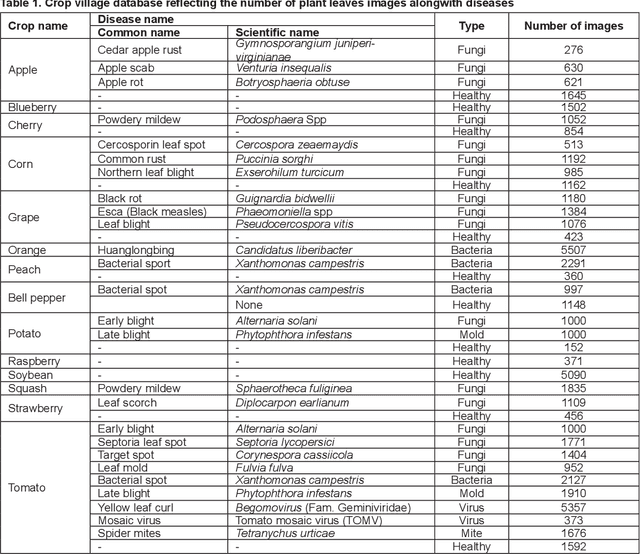
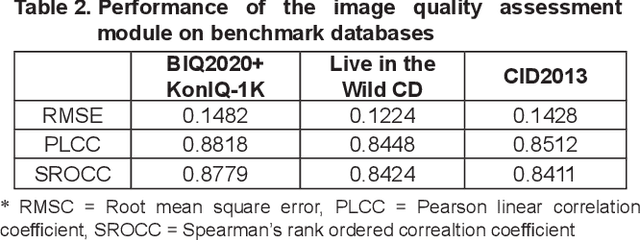
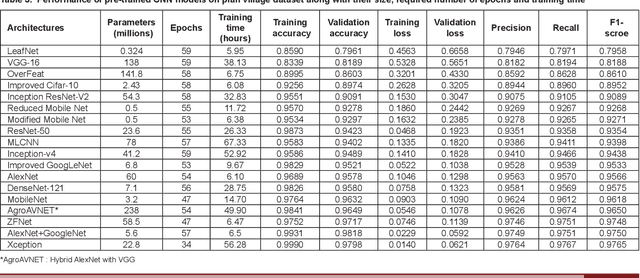
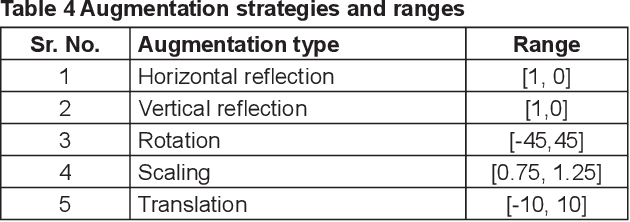
Abstract:Crop diseases are a major threat to food security and their rapid identification is important to prevent yield loss. Swift identification of these diseases are difficult due to the lack of necessary infrastructure. Recent advances in computer vision and increasing penetration of smartphones have paved the way for smartphone-assisted disease identification. Most of the plant diseases leave particular artifacts on the foliar structure of the plant. This study was conducted in 2020 at Department of Computer Science and Engineering, University of Engineering and Technology, Lahore, Pakistan to check leaf-based plant disease identification. This study provided a deep neural network-based solution to foliar disease identification and incorporated image quality assessment to select the image of the required quality to perform identification and named it Agricultural Pathologist (Agro Path). The captured image by a novice photographer may contain noise, lack of structure, and blur which result in a failed or inaccurate diagnosis. Moreover, AgroPath model had 99.42% accuracy for foliar disease identification. The proposed addition can be especially useful for application of foliar disease identification in the field of agriculture.
Text Mining Through Label Induction Grouping Algorithm Based Method
Dec 15, 2021
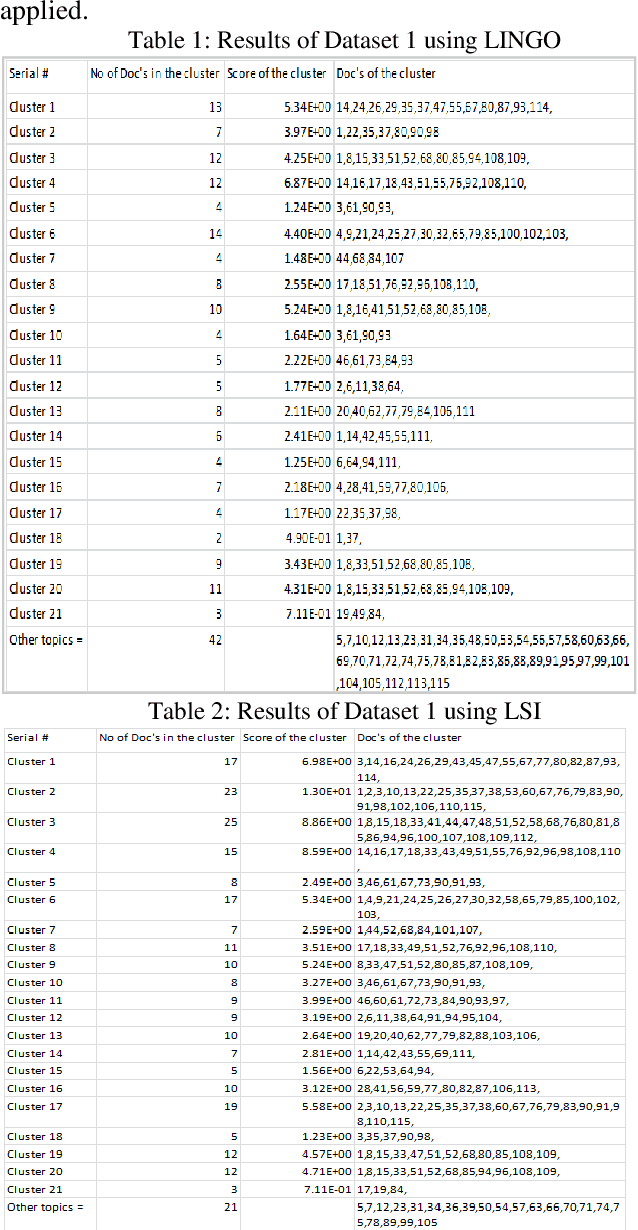
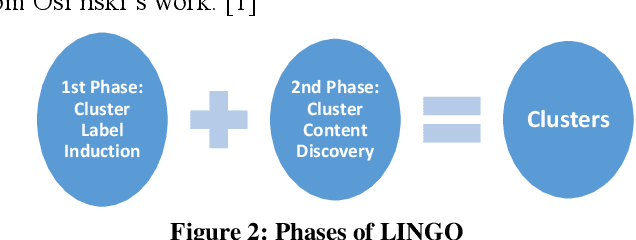

Abstract:The main focus of information retrieval methods is to provide accurate and efficient results which are cost-effective too. LINGO (Label Induction Grouping Algorithm) is a clustering algorithm that aims to provide search results in form of quality clusters but also has a few limitations. In this paper, our focus is based on achieving results that are more meaningful and improving the overall performance of the algorithm. LINGO works on two main steps; Cluster Label Induction by using Latent Semantic Indexing technique (LSI) and Cluster content discovery by using the Vector Space Model (VSM). As LINGO uses VSM in cluster content discovery, our task is to replace VSM with LSI for cluster content discovery and to analyze the feasibility of using LSI with Okapi BM25. The next task is to compare the results of a modified method with the LINGO original method. The research is applied to five different text-based data sets to get more reliable results for every method. Research results show that LINGO produces 40-50% better results when using LSI for content Discovery. From theoretical evidence using Okapi BM25 for scoring method in LSI (LSI+Okapi BM25) for cluster content discovery instead of VSM, also results in better clusters generation in terms of scalability and performance when compares to both VSM and LSI's Results.
* Presented in 5th International. Multidisciplinary Conference, 29-31 Oct., at, ICBS, Lahore
Development of Crop Yield Estimation Model using Soil and Environmental Parameters
Feb 10, 2021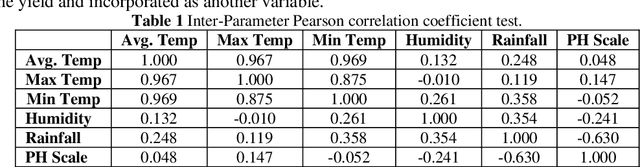
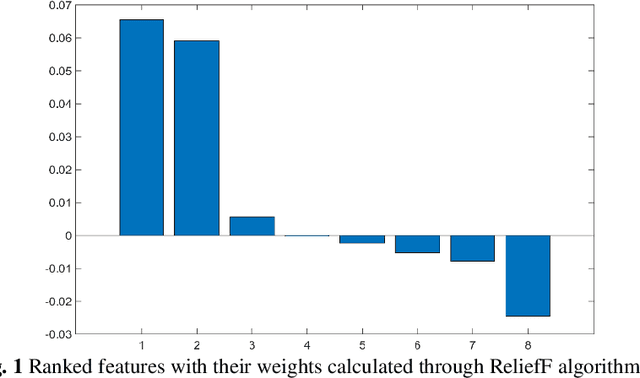
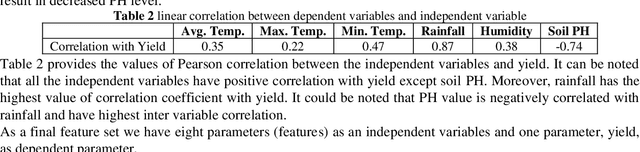
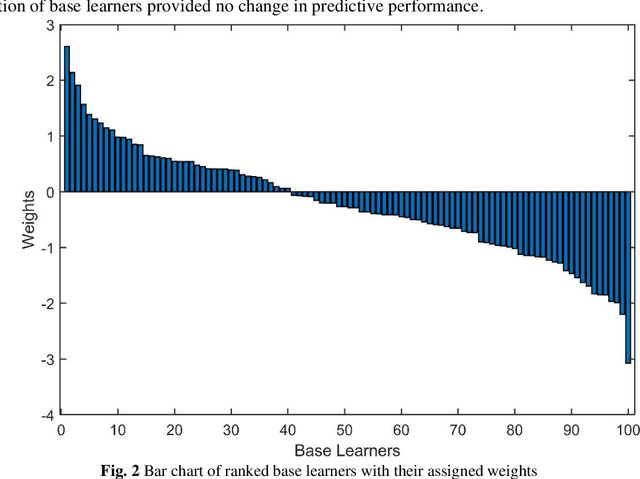
Abstract:Crop yield is affected by various soil and environmental parameters and can vary significantly. Therefore, a crop yield estimation model which can predict pre-harvest yield is required for food security. The study is conducted on tea forms operating under National Tea Research Institute, Pakistan. The data is recorded on monthly basis for ten years period. The parameters collected are minimum and maximum temperature, humidity, rainfall, PH level of the soil, usage of pesticide and labor expertise. The design of model incorporated all of these parameters and identified the parameters which are most crucial for yield predictions. Feature transformation is performed to obtain better performing model. The designed model is based on an ensemble of neural networks and provided an R-squared of 0.9461 and RMSE of 0.1204 indicating the usability of the proposed model in yield forecasting based on surface and environmental parameters.
* crop yield forecasting, regression, data mining, artificial neural network, ensemble learning
Leaf Image-based Plant Disease Identification using Color and Texture Features
Feb 08, 2021
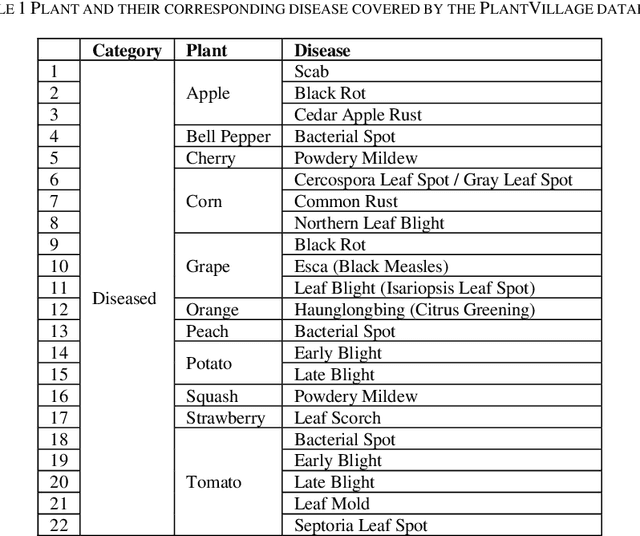
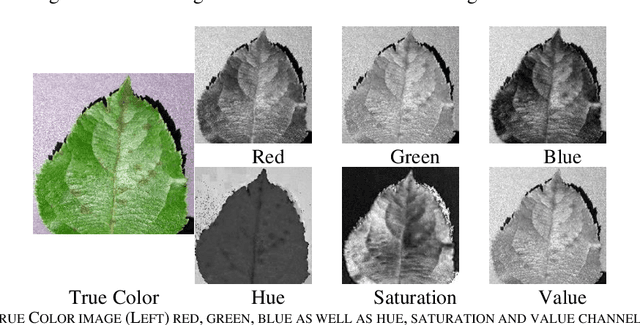
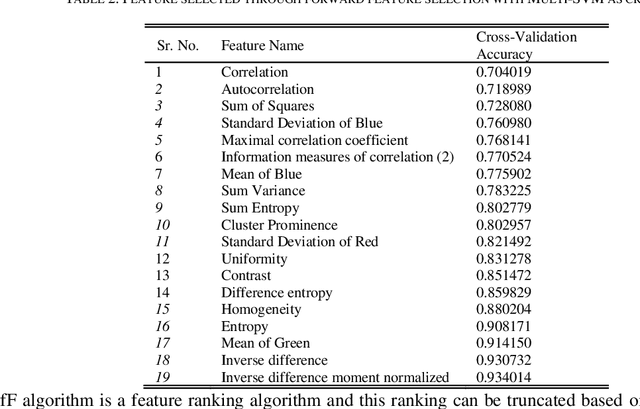
Abstract:Identification of plant disease is usually done through visual inspection or during laboratory examination which causes delays resulting in yield loss by the time identification is complete. On the other hand, complex deep learning models perform the task with reasonable performance but due to their large size and high computational requirements, they are not suited to mobile and handheld devices. Our proposed approach contributes automated identification of plant diseases which follows a sequence of steps involving pre-processing, segmentation of diseased leaf area, calculation of features based on the Gray-Level Co-occurrence Matrix (GLCM), feature selection and classification. In this study, six color features and twenty-two texture features have been calculated. Support vector machines is used to perform one-vs-one classification of plant disease. The proposed model of disease identification provides an accuracy of 98.79% with a standard deviation of 0.57 on 10-fold cross-validation. The accuracy on a self-collected dataset is 82.47% for disease identification and 91.40% for healthy and diseased classification. The reported performance measures are better or comparable to the existing approaches and highest among the feature-based methods, presenting it as the most suitable method to automated leaf-based plant disease identification. This prototype system can be extended by adding more disease categories or targeting specific crop or disease categories.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge