Guanghua He
MedVKAN: Efficient Feature Extraction with Mamba and KAN for Medical Image Segmentation
May 17, 2025Abstract:Medical image segmentation relies heavily on convolutional neural networks (CNNs) and Transformer-based models. However, CNNs are constrained by limited receptive fields, while Transformers suffer from scalability challenges due to their quadratic computational complexity. To address these limitations, recent advances have explored alternative architectures. The state-space model Mamba offers near-linear complexity while capturing long-range dependencies, and the Kolmogorov-Arnold Network (KAN) enhances nonlinear expressiveness by replacing fixed activation functions with learnable ones. Building on these strengths, we propose MedVKAN, an efficient feature extraction model integrating Mamba and KAN. Specifically, we introduce the EFC-KAN module, which enhances KAN with convolutional operations to improve local pixel interaction. We further design the VKAN module, integrating Mamba with EFC-KAN as a replacement for Transformer modules, significantly improving feature extraction. Extensive experiments on five public medical image segmentation datasets show that MedVKAN achieves state-of-the-art performance on four datasets and ranks second on the remaining one. These results validate the potential of Mamba and KAN for medical image segmentation while introducing an innovative and computationally efficient feature extraction framework. The code is available at: https://github.com/beginner-cjh/MedVKAN.
tCURLoRA: Tensor CUR Decomposition Based Low-Rank Parameter Adaptation for Medical Image Segmentation
Jan 04, 2025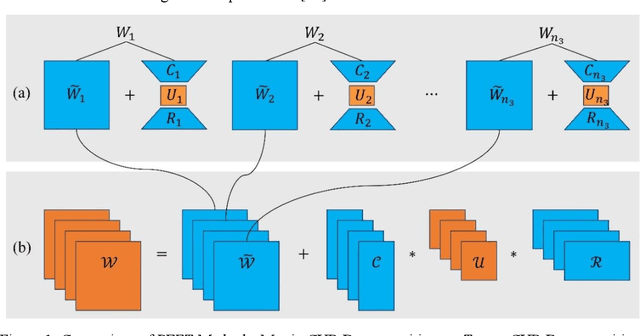

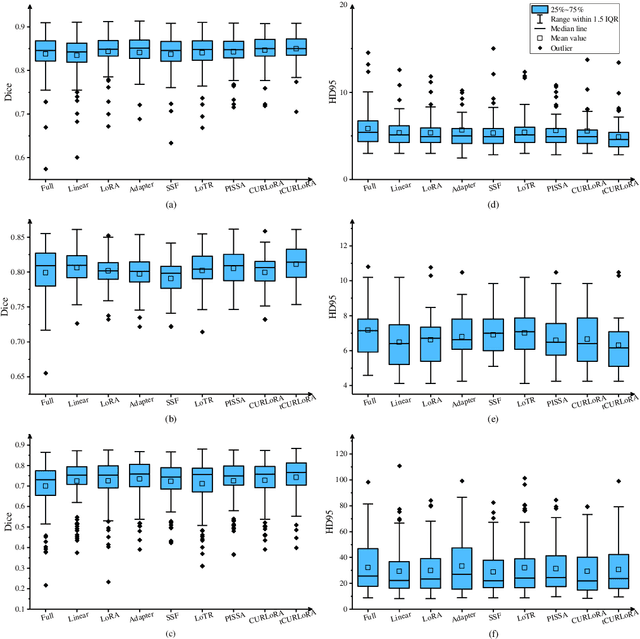
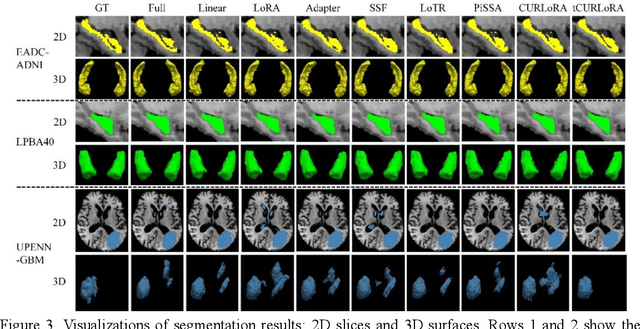
Abstract:Transfer learning, by leveraging knowledge from pre-trained models, has significantly enhanced the performance of target tasks. However, as deep neural networks scale up, full fine-tuning introduces substantial computational and storage challenges in resource-constrained environments, limiting its widespread adoption. To address this, parameter-efficient fine-tuning (PEFT) methods have been developed to reduce computational complexity and storage requirements by minimizing the number of updated parameters. While matrix decomposition-based PEFT methods, such as LoRA, show promise, they struggle to fully capture the high-dimensional structural characteristics of model weights. In contrast, high-dimensional tensors offer a more natural representation of neural network weights, allowing for a more comprehensive capture of higher-order features and multi-dimensional interactions. In this paper, we propose tCURLoRA, a novel fine-tuning method based on tensor CUR decomposition. By concatenating pre-trained weight matrices into a three-dimensional tensor and applying tensor CUR decomposition, we update only the lower-order tensor components during fine-tuning, effectively reducing computational and storage overhead. Experimental results demonstrate that tCURLoRA outperforms existing PEFT methods in medical image segmentation tasks.
A Novel Hybrid Parameter-Efficient Fine-Tuning Approach for Hippocampus Segmentation and Alzheimer's Disease Diagnosis
Sep 02, 2024



Abstract:Deep learning methods have significantly advanced medical image segmentation, yet their success hinges on large volumes of manually annotated data, which require specialized expertise for accurate labeling. Additionally, these methods often demand substantial computational resources, particularly for three-dimensional medical imaging tasks. Consequently, applying deep learning techniques for medical image segmentation with limited annotated data and computational resources remains a critical challenge. In this paper, we propose a novel parameter-efficient fine-tuning strategy, termed HyPS, which employs a hybrid parallel and serial architecture. HyPS updates a minimal subset of model parameters, thereby retaining the pre-trained model's original knowledge tructure while enhancing its ability to learn specific features relevant to downstream tasks. We apply this strategy to the state-of-the-art SwinUNETR model for medical image segmentation. Initially, the model is pre-trained on the BraTs2021 dataset, after which the HyPS method is employed to transfer it to three distinct hippocampus datasets.Extensive experiments demonstrate that HyPS outperforms baseline methods, especially in scenarios with limited training samples. Furthermore, based on the segmentation results, we calculated the hippocampal volumes of subjects from the ADNI dataset and combined these with metadata to classify disease types. In distinguishing Alzheimer's disease (AD) from cognitively normal (CN) individuals, as well as early mild cognitive impairment (EMCI) from late mild cognitive impairment (LMCI), HyPS achieved classification accuracies of 83.78% and 64.29%, respectively. These findings indicate that the HyPS method not only facilitates effective hippocampal segmentation using pre-trained models but also holds potential for aiding Alzheimer's disease detection. Our code is publicly available.
LoRA-PT: Low-Rank Adapting UNETR for Hippocampus Segmentation Using Principal Tensor Singular Values and Vectors
Jul 16, 2024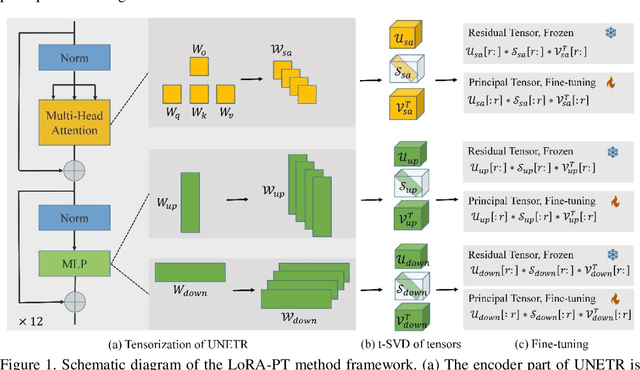
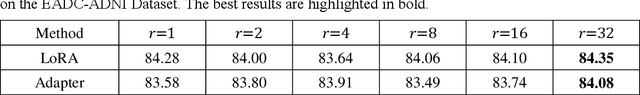
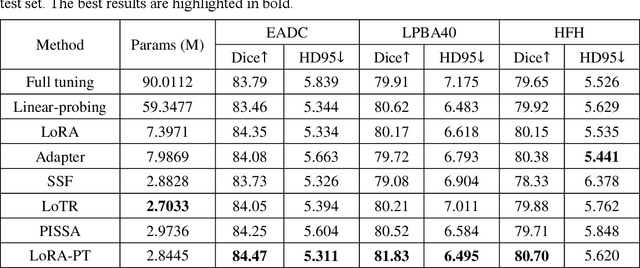
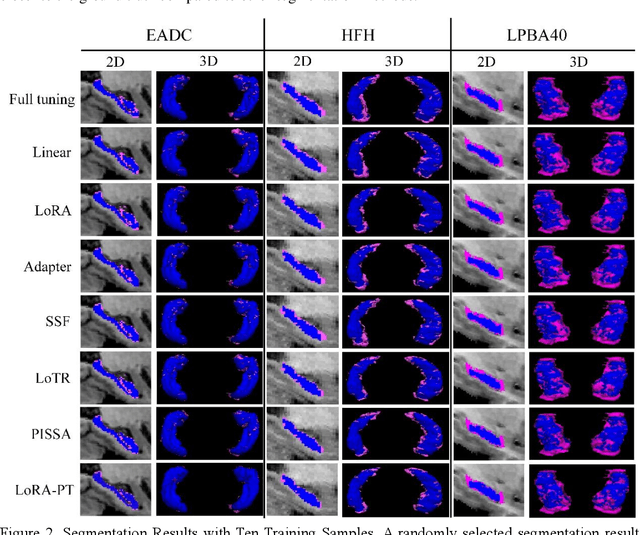
Abstract:The hippocampus is a crucial brain structure associated with various psychiatric disorders, and its automatic and precise segmentation is essential for studying these diseases. In recent years, deep learning-based methods have made significant progress in hippocampus segmentation. However, training deep neural network models requires substantial computational resources and time, as well as a large amount of labeled training data, which is often difficult to obtain in medical image segmentation. To address this issue, we propose a new parameter-efficient fine-tuning method called LoRA-PT. This method transfers the pre-trained UNETR model on the BraTS2021 dataset to the hippocampus segmentation task. Specifically, the LoRA-PT method categorizes the parameter matrix of the transformer structure into three sizes, forming three 3D tensors. Through tensor singular value decomposition, these tensors are decomposed to generate low-rank tensors with the principal singular values and singular vectors, while the remaining singular values and vectors form the residual tensor. Similar to the LoRA method, during parameter fine-tuning, we only update the low-rank tensors, i.e. the principal tensor singular values and vectors, while keeping the residual tensor unchanged. We validated the proposed method on three public hippocampus datasets. Experimental results show that LoRA-PT outperforms existing parameter-efficient transfer learning methods in segmentation accuracy while significantly reducing the number of parameter updates.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge