Georg Hille
Joint Liver and Hepatic Lesion Segmentation using a Hybrid CNN with Transformer Layers
Jan 26, 2022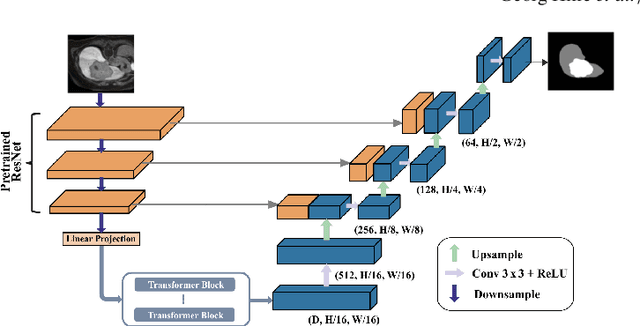
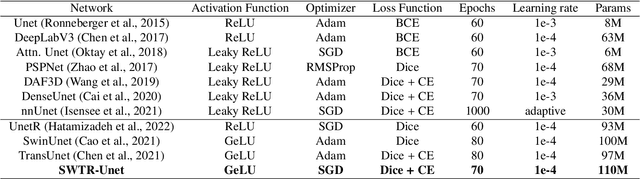
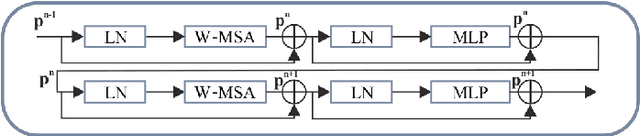
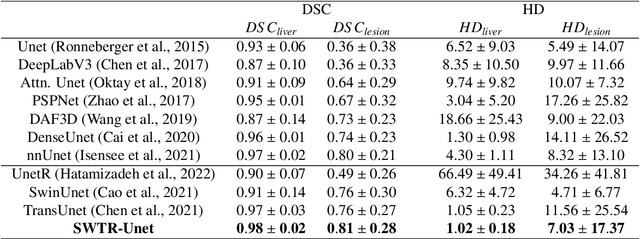
Abstract:Deep learning-based segmentation of the liver and hepatic lesions therein steadily gains relevance in clinical practice due to the increasing incidence of liver cancer each year. Whereas various network variants with overall promising results in the field of medical image segmentation have been developed over the last years, almost all of them struggle with the challenge of accurately segmenting hepatic lesions. This lead to the idea of combining elements of convolutional and transformerbased architectures to overcome the existing limitations. This work presents a hybrid network called SWTR-Unet, consisting of a pretrained ResNet, transformer blocks as well as a common Unet-style decoder path. This network was applied to clinical liver MRI, as well as to the publicly available CT data of the liver tumor segmentation (LiTS) challenge. Additionally, multiple state-of-the-art networks were implemented and applied to both datasets, ensuring a direct comparability. Furthermore, correlation analysis and an ablation study were carried out, to investigate various influencing factors on the segmentation accuracy of our presented method. With Dice similarity scores of averaged 98 +- 2 % for liver and 81 +- 28 % lesion segmentation on the MRI dataset and 97 +- 2 % and 79 +- 25 %, respectively on the CT dataset, the proposed SWTR-Unet outperforms each of the additionally implemented state-of-the-art networks. The achieved segmentation accuracy was found to be on par with manually performed expert segmentations as indicated by interobserver variabilities for liver lesion segmentation. In conclusion, the presented method could save valuable time and resources in clinical practice.
Spinal Metastases Segmentation in MR Imaging using Deep Convolutional Neural Networks
Jan 28, 2020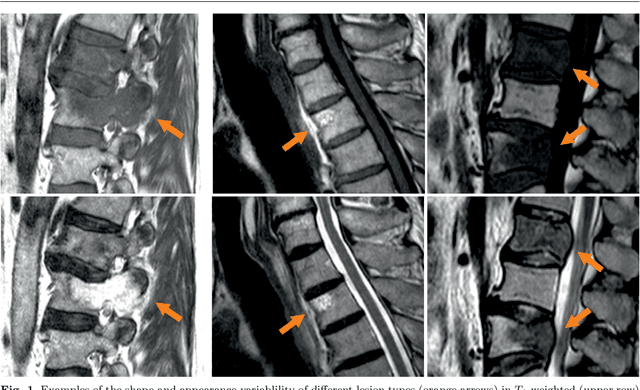
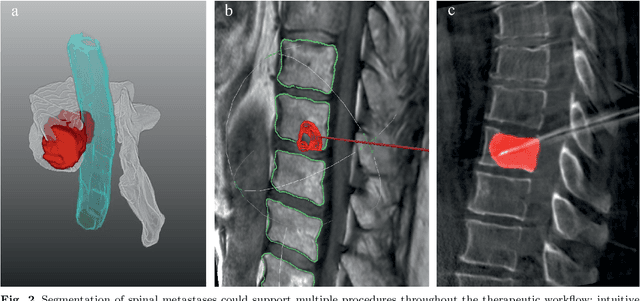
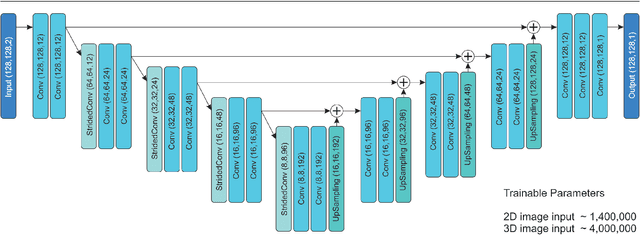
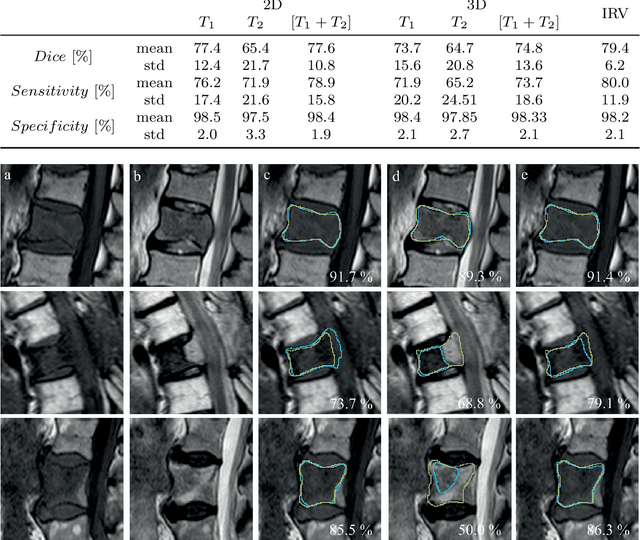
Abstract:This study's objective was to segment spinal metastases in diagnostic MR images using a deep learning-based approach. Segmentation of such lesions can present a pivotal step towards enhanced therapy planning and validation, as well as intervention support during minimally invasive and image-guided surgeries like radiofrequency ablations. For this purpose, we used a U-Net like architecture trained with 40 clinical cases including both, lytic and sclerotic lesion types and various MR sequences. Our proposed method was evaluated with regards to various factors influencing the segmentation quality, e.g. the used MR sequences and the input dimension. We quantitatively assessed our experiments using Dice coefficients, sensitivity and specificity rates. Compared to expertly annotated lesion segmentations, the experiments yielded promising results with average Dice scores up to 77.6% and mean sensitivity rates up to 78.9%. To our best knowledge, our proposed study is one of the first to tackle this particular issue, which limits direct comparability with related works. In respect to similar deep learning-based lesion segmentations, e.g. in liver MR images or spinal CT images, our experiments showed similar or in some respects superior segmentation quality. Overall, our automatic approach can provide almost expert-like segmentation accuracy in this challenging and ambitious task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge