Gentry Atkinson
BioDiffusion: A Versatile Diffusion Model for Biomedical Signal Synthesis
Jan 27, 2024



Abstract:Machine learning tasks involving biomedical signals frequently grapple with issues such as limited data availability, imbalanced datasets, labeling complexities, and the interference of measurement noise. These challenges often hinder the optimal training of machine learning algorithms. Addressing these concerns, we introduce BioDiffusion, a diffusion-based probabilistic model optimized for the synthesis of multivariate biomedical signals. BioDiffusion demonstrates excellence in producing high-fidelity, non-stationary, multivariate signals for a range of tasks including unconditional, label-conditional, and signal-conditional generation. Leveraging these synthesized signals offers a notable solution to the aforementioned challenges. Our research encompasses both qualitative and quantitative assessments of the synthesized data quality, underscoring its capacity to bolster accuracy in machine learning tasks tied to biomedical signals. Furthermore, when juxtaposed with current leading time-series generative models, empirical evidence suggests that BioDiffusion outperforms them in biomedical signal generation quality.
Measure Twice, Cut Once: Quantifying Bias and Fairness in Deep Neural Networks
Oct 08, 2021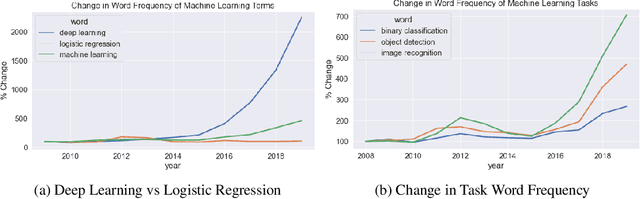
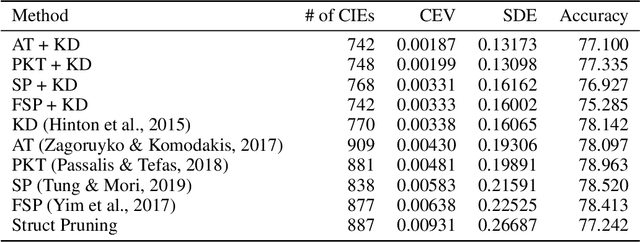
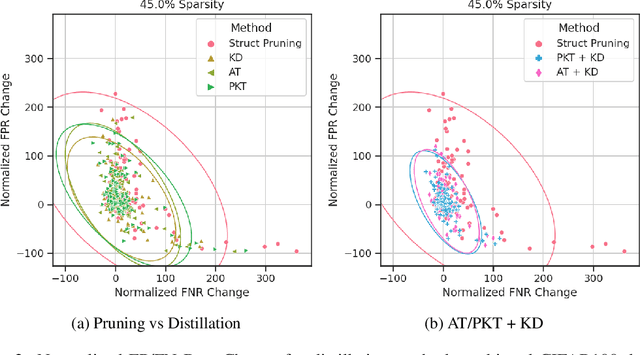
Abstract:Algorithmic bias is of increasing concern, both to the research community, and society at large. Bias in AI is more abstract and unintuitive than traditional forms of discrimination and can be more difficult to detect and mitigate. A clear gap exists in the current literature on evaluating the relative bias in the performance of multi-class classifiers. In this work, we propose two simple yet effective metrics, Combined Error Variance (CEV) and Symmetric Distance Error (SDE), to quantitatively evaluate the class-wise bias of two models in comparison to one another. By evaluating the performance of these new metrics and by demonstrating their practical application, we show that they can be used to measure fairness as well as bias. These demonstrations show that our metrics can address specific needs for measuring bias in multi-class classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge