Vangelis Metsis
DyWPE: Signal-Aware Dynamic Wavelet Positional Encoding for Time Series Transformers
Sep 18, 2025Abstract:Existing positional encoding methods in transformers are fundamentally signal-agnostic, deriving positional information solely from sequence indices while ignoring the underlying signal characteristics. This limitation is particularly problematic for time series analysis, where signals exhibit complex, non-stationary dynamics across multiple temporal scales. We introduce Dynamic Wavelet Positional Encoding (DyWPE), a novel signal-aware framework that generates positional embeddings directly from input time series using the Discrete Wavelet Transform (DWT). Comprehensive experiments in ten diverse time series datasets demonstrate that DyWPE consistently outperforms eight existing state-of-the-art positional encoding methods, achieving average relative improvements of 9.1\% compared to baseline sinusoidal absolute position encoding in biomedical signals, while maintaining competitive computational efficiency.
A Systematic Evaluation of LLM Strategies for Mental Health Text Analysis: Fine-tuning vs. Prompt Engineering vs. RAG
Mar 31, 2025Abstract:This study presents a systematic comparison of three approaches for the analysis of mental health text using large language models (LLMs): prompt engineering, retrieval augmented generation (RAG), and fine-tuning. Using LLaMA 3, we evaluate these approaches on emotion classification and mental health condition detection tasks across two datasets. Fine-tuning achieves the highest accuracy (91% for emotion classification, 80% for mental health conditions) but requires substantial computational resources and large training sets, while prompt engineering and RAG offer more flexible deployment with moderate performance (40-68% accuracy). Our findings provide practical insights for implementing LLM-based solutions in mental health applications, highlighting the trade-offs between accuracy, computational requirements, and deployment flexibility.
Positional Encoding in Transformer-Based Time Series Models: A Survey
Feb 17, 2025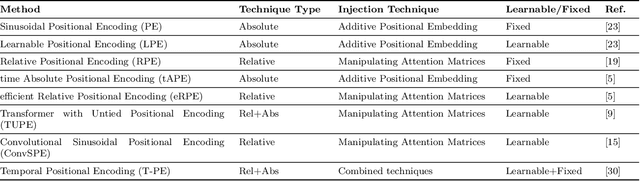
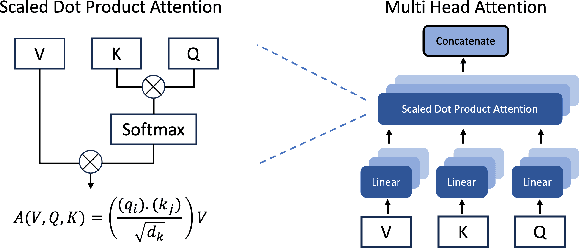
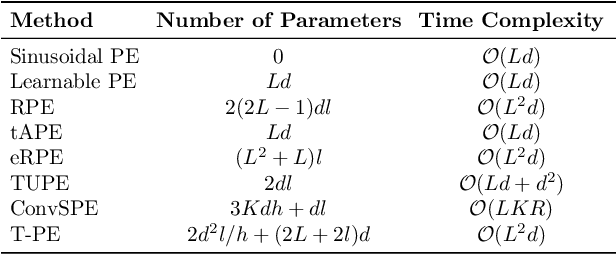
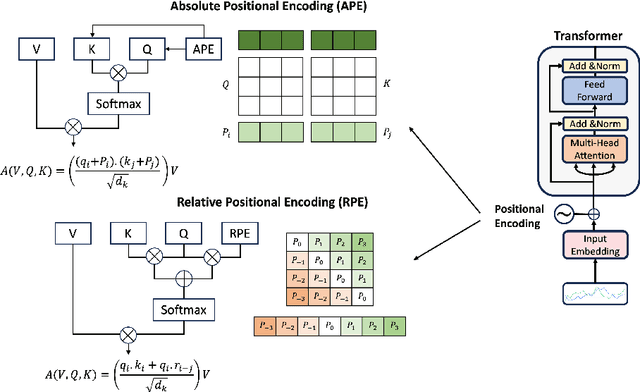
Abstract:Recent advancements in transformer-based models have greatly improved time series analysis, providing robust solutions for tasks such as forecasting, anomaly detection, and classification. A crucial element of these models is positional encoding, which allows transformers to capture the intrinsic sequential nature of time series data. This survey systematically examines existing techniques for positional encoding in transformer-based time series models. We investigate a variety of methods, including fixed, learnable, relative, and hybrid approaches, and evaluate their effectiveness in different time series classification tasks. Furthermore, we outline key challenges and suggest potential research directions to enhance positional encoding strategies. By delivering a comprehensive overview and quantitative benchmarking, this survey intends to assist researchers and practitioners in selecting and designing effective positional encoding methods for transformer-based time series models.
BioDiffusion: A Versatile Diffusion Model for Biomedical Signal Synthesis
Jan 27, 2024



Abstract:Machine learning tasks involving biomedical signals frequently grapple with issues such as limited data availability, imbalanced datasets, labeling complexities, and the interference of measurement noise. These challenges often hinder the optimal training of machine learning algorithms. Addressing these concerns, we introduce BioDiffusion, a diffusion-based probabilistic model optimized for the synthesis of multivariate biomedical signals. BioDiffusion demonstrates excellence in producing high-fidelity, non-stationary, multivariate signals for a range of tasks including unconditional, label-conditional, and signal-conditional generation. Leveraging these synthesized signals offers a notable solution to the aforementioned challenges. Our research encompasses both qualitative and quantitative assessments of the synthesized data quality, underscoring its capacity to bolster accuracy in machine learning tasks tied to biomedical signals. Furthermore, when juxtaposed with current leading time-series generative models, empirical evidence suggests that BioDiffusion outperforms them in biomedical signal generation quality.
TTS-CGAN: A Transformer Time-Series Conditional GAN for Biosignal Data Augmentation
Jun 28, 2022
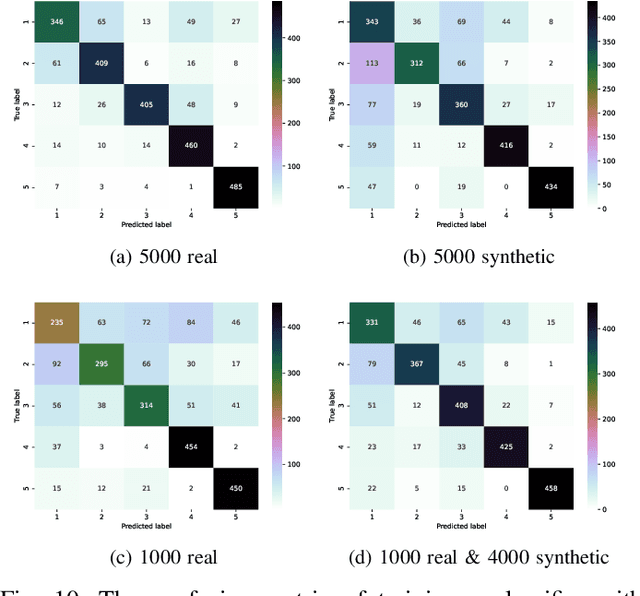
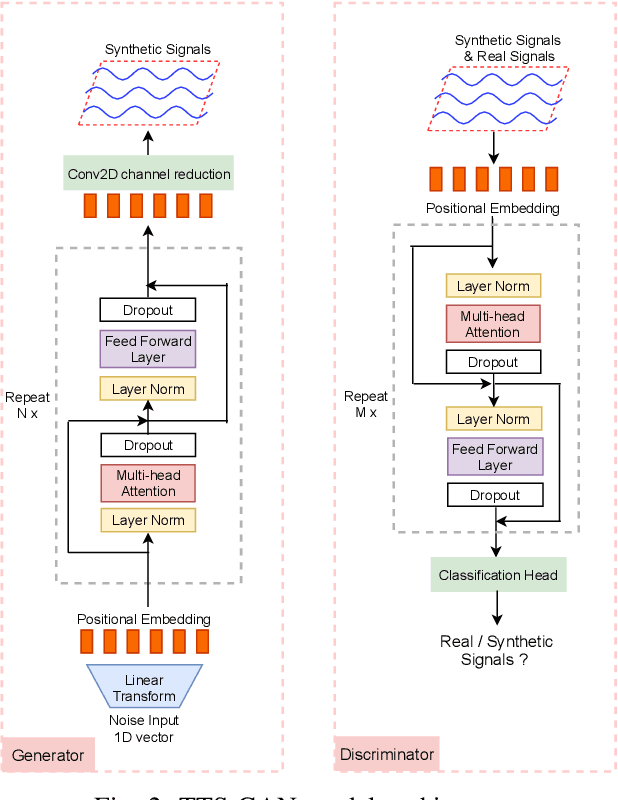
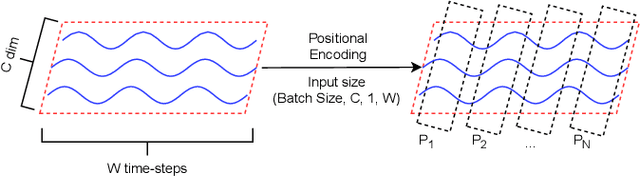
Abstract:Signal measurement appearing in the form of time series is one of the most common types of data used in medical machine learning applications. Such datasets are often small in size, expensive to collect and annotate, and might involve privacy issues, which hinders our ability to train large, state-of-the-art deep learning models for biomedical applications. For time-series data, the suite of data augmentation strategies we can use to expand the size of the dataset is limited by the need to maintain the basic properties of the signal. Generative Adversarial Networks (GANs) can be utilized as another data augmentation tool. In this paper, we present TTS-CGAN, a transformer-based conditional GAN model that can be trained on existing multi-class datasets and generate class-specific synthetic time-series sequences of arbitrary length. We elaborate on the model architecture and design strategies. Synthetic sequences generated by our model are indistinguishable from real ones, and can be used to complement or replace real signals of the same type, thus achieving the goal of data augmentation. To evaluate the quality of the generated data, we modify the wavelet coherence metric to be able to compare the similarity between two sets of signals, and also conduct a case study where a mix of synthetic and real data are used to train a deep learning model for sequence classification. Together with other visualization techniques and qualitative evaluation approaches, we demonstrate that TTS-CGAN generated synthetic data are similar to real data, and that our model performs better than the other state-of-the-art GAN models built for time-series data generation.
TTS-GAN: A Transformer-based Time-Series Generative Adversarial Network
Feb 06, 2022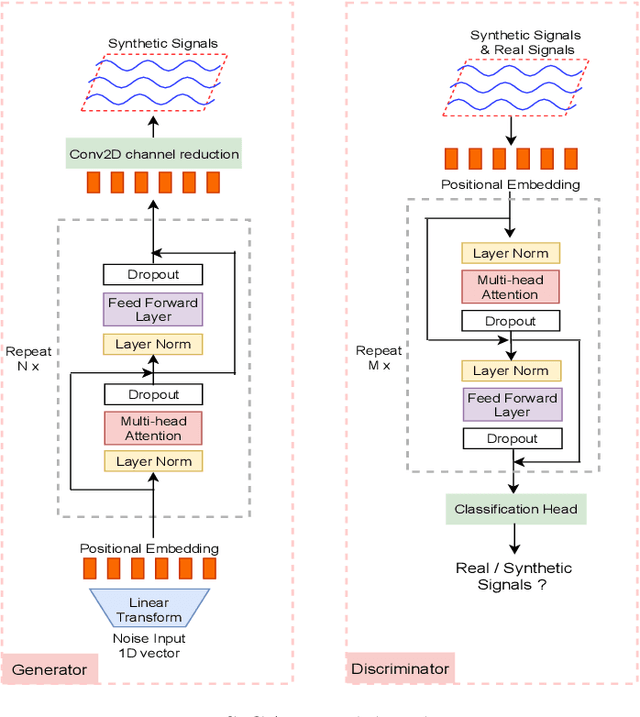
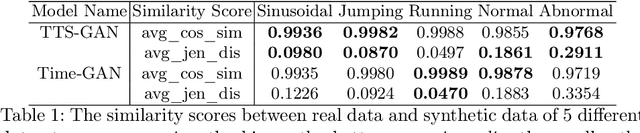
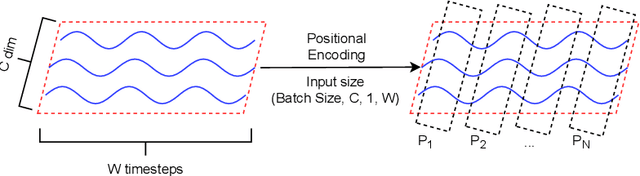
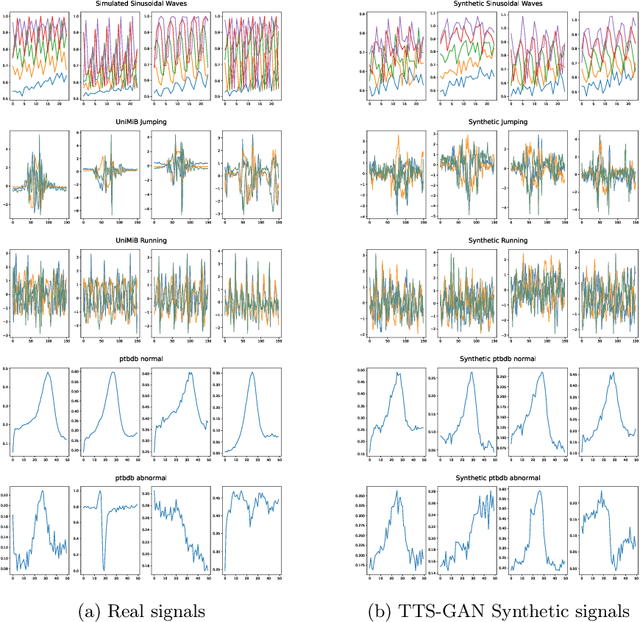
Abstract:Signal measurements appearing in the form of time series are one of the most common types of data used in medical machine learning applications. However, such datasets are often small, making the training of deep neural network architectures ineffective. For time-series, the suite of data augmentation tricks we can use to expand the size of the dataset is limited by the need to maintain the basic properties of the signal. Data generated by a Generative Adversarial Network (GAN) can be utilized as another data augmentation tool. RNN-based GANs suffer from the fact that they cannot effectively model long sequences of data points with irregular temporal relations. To tackle these problems, we introduce TTS-GAN, a transformer-based GAN which can successfully generate realistic synthetic time-series data sequences of arbitrary length, similar to the real ones. Both the generator and discriminator networks of the GAN model are built using a pure transformer encoder architecture. We use visualizations and dimensionality reduction techniques to demonstrate the similarity of real and generated time-series data. We also compare the quality of our generated data with the best existing alternative, which is an RNN-based time-series GAN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge