Frederic Runge
Towards Automated Design of Riboswitches
Jul 17, 2023



Abstract:Experimental screening and selection pipelines for the discovery of novel riboswitches are expensive, time-consuming, and inefficient. Using computational methods to reduce the number of candidates for the screen could drastically decrease these costs. However, existing computational approaches do not fully satisfy all requirements for the design of such initial screening libraries. In this work, we present a new method, libLEARNA, capable of providing RNA focus libraries of diverse variable-length qualified candidates. Our novel structure-based design approach considers global properties as well as desired sequence and structure features. We demonstrate the benefits of our method by designing theophylline riboswitch libraries, following a previously published protocol, and yielding 30% more unique high-quality candidates.
Scalable Deep Learning for RNA Secondary Structure Prediction
Jul 14, 2023



Abstract:The field of RNA secondary structure prediction has made significant progress with the adoption of deep learning techniques. In this work, we present the RNAformer, a lean deep learning model using axial attention and recycling in the latent space. We gain performance improvements by designing the architecture for modeling the adjacency matrix directly in the latent space and by scaling the size of the model. Our approach achieves state-of-the-art performance on the popular TS0 benchmark dataset and even outperforms methods that use external information. Further, we show experimentally that the RNAformer can learn a biophysical model of the RNA folding process.
Probabilistic Transformer: Modelling Ambiguities and Distributions for RNA Folding and Molecule Design
May 27, 2022



Abstract:Our world is ambiguous and this is reflected in the data we use to train our algorithms. This is especially true when we try to model natural processes where collected data is affected by noisy measurements and differences in measurement techniques. Sometimes, the process itself can be ambiguous, such as in the case of RNA folding, where a single nucleotide sequence can fold into multiple structures. This ambiguity suggests that a predictive model should have similar probabilistic characteristics to match the data it models. Therefore, we propose a hierarchical latent distribution to enhance one of the most successful deep learning models, the Transformer, to accommodate ambiguities and data distributions. We show the benefits of our approach on a synthetic task, with state-of-the-art results in RNA folding, and demonstrate its generative capabilities on property-based molecule design, outperforming existing work.
Learning to Design RNA
Dec 31, 2018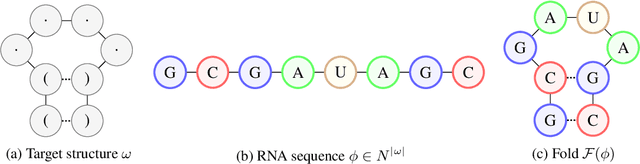
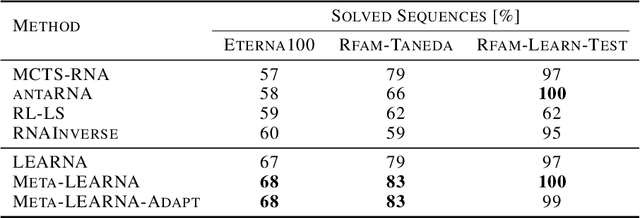
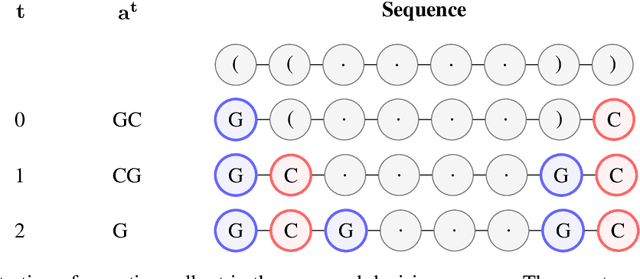
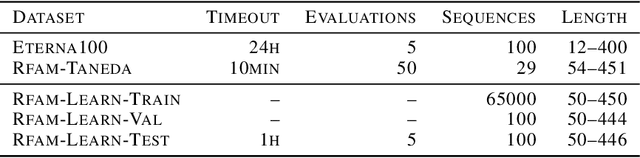
Abstract:Designing RNA molecules has garnered recent interest in medicine, synthetic biology, biotechnology and bioinformatics since many functional RNA molecules were shown to be involved in regulatory processes for transcription, epigenetics and translation. Since an RNA's function depends on its structural properties, the RNA Design problem is to find an RNA sequence that folds into a specified secondary structure. Here, we propose a new algorithm for the RNA Design problem, dubbed LEARNA. LEARNA uses deep reinforcement learning to train a policy network to sequentially design an entire RNA sequence given a specified secondary target structure. By meta-learning across 8000 different RNA target structures for one hour on 20 cores, our extension Meta-LEARNA constructs an RNA Design policy that can be applied out of the box to solve novel RNA target structures. Methodologically, for what we believe to be the first time, we jointly optimize over a rich space of neural architectures for the policy network, the hyperparameters of the training procedure and the formulation of the decision process. Comprehensive empirical results on two widely-used RNA secondary structure design benchmarks, as well as a third one that we introduce, show that our approach achieves new state-of-the-art performance on all benchmarks while also being orders of magnitudes faster in reaching the previous state-of-the-art performance. In an ablation study, we analyze the importance of our method's different components.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge