Frank M Sanfilippo
Explainable Artificial Intelligence for Pharmacovigilance: What Features Are Important When Predicting Adverse Outcomes?
Dec 25, 2021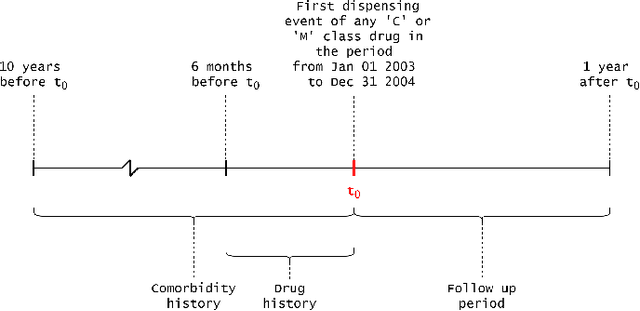
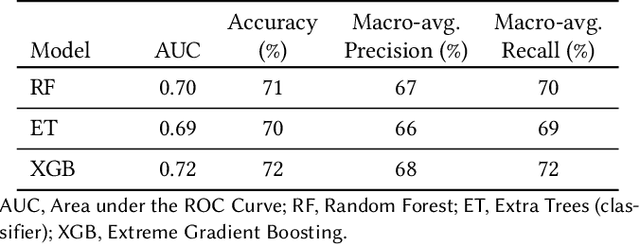
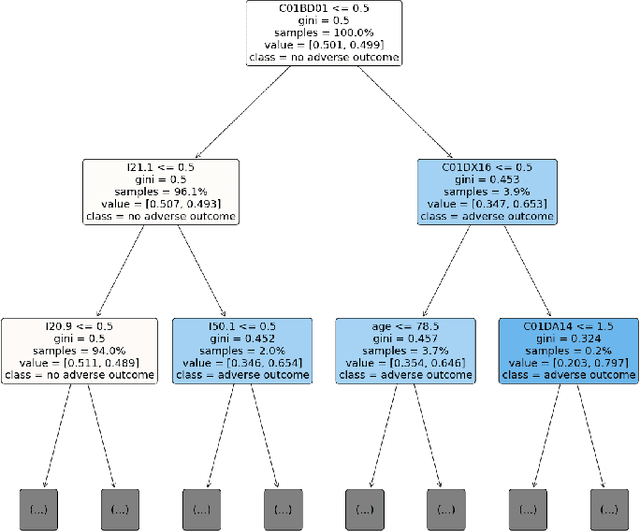
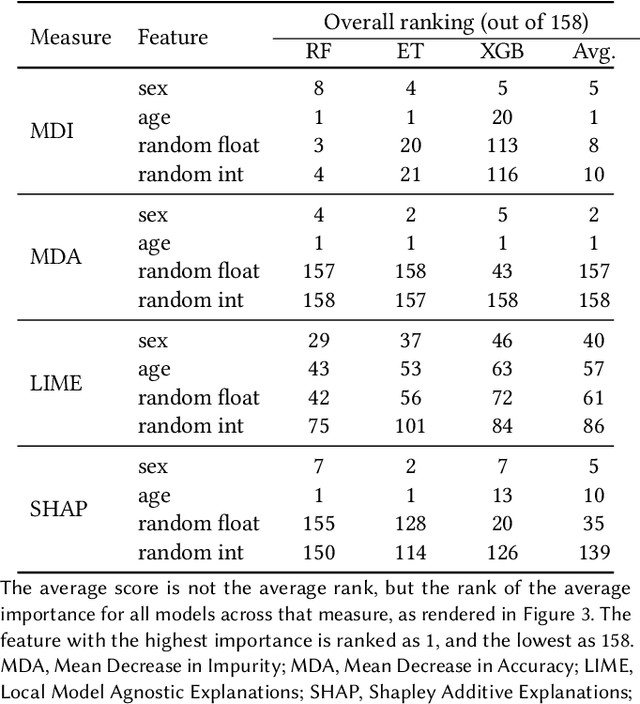
Abstract:Explainable Artificial Intelligence (XAI) has been identified as a viable method for determining the importance of features when making predictions using Machine Learning (ML) models. In this study, we created models that take an individual's health information (e.g. their drug history and comorbidities) as inputs, and predict the probability that the individual will have an Acute Coronary Syndrome (ACS) adverse outcome. Using XAI, we quantified the contribution that specific drugs had on these ACS predictions, thus creating an XAI-based technique for pharmacovigilance monitoring, using ACS as an example of the adverse outcome to detect. Individuals aged over 65 who were supplied Musculo-skeletal system (anatomical therapeutic chemical (ATC) class M) or Cardiovascular system (ATC class C) drugs between 1993 and 2009 were identified, and their drug histories, comorbidities, and other key features were extracted from linked Western Australian datasets. Multiple ML models were trained to predict if these individuals would have an ACS related adverse outcome (i.e., death or hospitalisation with a discharge diagnosis of ACS), and a variety of ML and XAI techniques were used to calculate which features -- specifically which drugs -- led to these predictions. The drug dispensing features for rofecoxib and celecoxib were found to have a greater than zero contribution to ACS related adverse outcome predictions (on average), and it was found that ACS related adverse outcomes can be predicted with 72% accuracy. Furthermore, the XAI libraries LIME and SHAP were found to successfully identify both important and unimportant features, with SHAP slightly outperforming LIME. ML models trained on linked administrative health datasets in tandem with XAI algorithms can successfully quantify feature importance, and with further development, could potentially be used as pharmacovigilance monitoring techniques.
Imputation of Missing Data with Class Imbalance using Conditional Generative Adversarial Networks
Dec 01, 2020



Abstract:Missing data is a common problem faced with real-world datasets. Imputation is a widely used technique to estimate the missing data. State-of-the-art imputation approaches, such as Generative Adversarial Imputation Nets (GAIN), model the distribution of observed data to approximate the missing values. Such an approach usually models a single distribution for the entire dataset, which overlooks the class-specific characteristics of the data. Class-specific characteristics are especially useful when there is a class imbalance. We propose a new method for imputing missing data based on its class-specific characteristics by adapting the popular Conditional Generative Adversarial Networks (CGAN). Our Conditional Generative Adversarial Imputation Network (CGAIN) imputes the missing data using class-specific distributions, which can produce the best estimates for the missing values. We tested our approach on benchmark datasets and achieved superior performance compared with the state-of-the-art and popular imputation approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge